NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
It is becoming clear that in prokaryotes RNAs interact and perform complex functions as a network similar to what we have uncovered in eukaryotes. This chapter will continue the discussion of prokaryotic molecular systems, showing how these systems can interact with each other to gain a higher level of control within the cell. Our examples include RNase P, the tRNA cleaving molecule that, as well as performing other functions, also cleaves certain riboswitches; and the glmS gene under the control of both a ribozyme in its 5′ untranslated region and two small RNAs. With further investigation of nonprotein coding RNA interactions (i.e., the RNA infrastructure), in bacteria and archaea, we gain greater understanding of the influence that small strands of RNA sequence can have over the entire cell.
INTRODUCTION
The previous chapter began our discussion of the RNA networks within the prokaryotic cell with an overview of the CRISPR system of viral defence, and riboswitch regulation. Although both of these systems have equivalents within eukaryotes, it is clear that prokaryotes operate these mechanisms in their own way. In a similar way, this chapter will look at some further RNA-based networks in the prokaryotic cell, some of which are unique to prokaryotes and some with similar networks in eukaryotes.
A key endoribonuclease RNase P is examined first in relation to its role in RNA cleavage and the removal of leader sequences, and since it potentially affects many hundreds of genes, it can be considered central to a large bacterial RNA network. Small RNAs and sigma factors are discussed next, with a focus on a key bacterial regulator, the sigma factor RpoS and its role in sensing the cellular environment and detecting stress. Finally, a relatively newcomer to bacterial RNA network studies, transfer messenger RNA, a cross between tRNA and mRNA responsible for releasing stalled ribosomes, is discussed in light of its role in translation and bacterial sporulation. With new studies on prokaryotic RNAs being released almost daily, it is impossible to cover all the angles surrounding RNA networks. These examples and those from the previous chapter therefore, give a taste of what is likely to become in the near future, a complex RNA-infrastructure similar to that seen in eukaryotes, but with its own unique flavour.
RNase P AND tRNA PROCESSING IN BACTERIA AND ARCHAEA
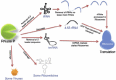
RNase P is a site-specific endoribonuclease which plays a central role in the prokaryotic RNA-processing network (Fig. 1), (for a review see refs. 1-4). In addition to its primary action of cleaving pre-tRNAs to progress their maturation, RNase P has been shown to cleave other substrates in bacteria such as pre-4.5S RNA, pre-tmRNA, polycistronic operons, viral transcripts and riboswitches. Although present in all three kingdoms of life (i.e., eukaryotes, bacteria and archaea) the RNase P holoenzyme has a very different macromolecular structure in each kingdom, and there are differences in how RNase P interacts with other RNAs in each kingdom's RNA infrastructure.
In bacteria, RNase P consists of a single RNA (mass ~120 kDa) and a single protein (mass ~15 kDa).2 Although this basic protein is small, it plays diverse roles such as enhancing substrate binding, altering substrate recognition, stabilising RNA conformation, and aiding catalysis by discriminating between the substrate and product by binding to the 5′ leader sequence of the pre-tRNA.1,2 In archaea multiple proteins (five including the ribosomal protein L7Ae5),bind to the single RNA.6 These archaeal proteins show some evolutionary relationship to eukaryotic RNase P proteins and most were identified by sequence similarity to yeast proteins (summarised in ref. 7). However, the Mth687 (pop5) protein from Methanothermobacter thermoautotrophicus adopts a fold similar to that of the bacterial protein and this protein may carry out some of the functions found in the bacterial protein.3,7 The RNase P RNA from some representatives from each kingdom can be induced to perform weak catalysis without its accompanying proteins, but only with high salt and high cation conditions in vitro (summarised in ref. 7).
Within the bacterial RNase P holoenzyme, the protein and RNA subunits function synergistically in both substrate recognition and catalysis, to process the products of more than 80 pre-tRNA genes (Escherichia coli and Bacillus subtilis).8 In all cases the action of RNase P is straightforward, it cleaves sequences at a designated site and most interactions are RNA-RNA between the RNase P RNA and the substrate RNA.
Outside of cleaving 5′ leader sequences of tRNAs and tmRNAs (discussed later) there are a number of other targets for RNase P cleavage. Some polycistronic mRNA transcripts can be processed with RNase P in E. coli independent of RNase E (which is typically required).9 For example, with the valV-valW and LeuQ-leuP-leuV transcripts only the RNase P activity is necessary to generate pre-tRNAs with mature 5′ ends.9 The 4.5S RNA substrate which is part of the Signal Recognition Particle (SRP) responsible for post-translational transport, is also cleaved by RNase P. Interestingly the structure of the 4.5S RNA is very different from the tRNA clover-leaf shape, forming instead a long hairpin. The phage Φ80-induced RNA has a similar hairpin structure and is also cleaved by RNase P (reviewed in ref. 3). In other viral examples, the antisense RNA precursor C4 from bacteriophages P1 and P7 is cleaved by RNase P resulting in the inhibition of Ant (antirepressor) synthesis.3 tRNA-like structures from the turnip yellow mosaic virus (RYMV) can also be cleaved by RNase P RNA from E. coli,10 thus leading to research in using RNase P as an antiviral agent for animals.11
RNase P has also been shown to cleave the 5′ regions of some riboswitches, including the adenine riboswitch from B. subtilis12 and co-enzyme B12 riboswitches (btuB) from E. coli and B. subtilis.13 RNase P cleavage of riboswitches is not a general mechanism, as not all riboswitches are cleaved by RNase P (e.g., TPP, FMN, SAM and lysine riboswitches of E. coli13 and the xpt-pbuX guanine riboswitch of B. subtilis12 are not cleaved). The riboswitches that are cleaved do not display a tRNA-like cleavage site or even a model cleavage site.13 The B. subtilis btuB riboswitch cleavage site is located in a region that has a short single-stranded region next to a putative double-stranded region but such a structural motif was not found in the E. coli btuB riboswitch.13 The pbuE gene is controlled by the adenine riboswitch and in the absence of adenine, the riboswitch folds into an 'off' structure which includes a transcription terminator. The pbuE adenine riboswitch is cleaved at two sites C (-27) and C (-138), with cleavage efficiency of the latter dependent on the length of the substrate, with shorter substrates displaying decreased efficiency.12
BACTERIAL SMALL RNAs AND SIGMA FACTORS
In bacteria small RNAs (sRNAs) typically ~100 nt long have been identified (~100 of them in E. coli), mostly in intergenic regions flanked by recognisable promoters and/or Rho-independent terminators14 (reviewed in refs. 15, 16). Studied mainly in E. coli and Salmonella, they have been associated with many metabolic pathways including stress responses to oxygen levels17 and drugs,18 carbohydrate uptake and metabolism,19 iron metabolism20 and quorum sensing,21 and act to regulate gene expression post-transcriptionally. Some sRNAs such as Spot42 have been known since the early 1970s as an abundant RNA species,19 whereas others are just coming to light as the roles of small RNAs in bacterial mechanisms is more widely studied. Systems biology approaches using bacterialike E. coli have allowed for the role of some sRNAs within the transcriptional regulatory network to be examined (reviewed within ref. 18). Furthermore, because of their small size and lack of translation, sRNAs have been hypothesised to be cost effective to the cell from a regulation point of view. Mathematical modelling of the sRNA-mRNA interaction has shown that this is a good mechanism when quick responses to external stimuli are required, consistent with experimental results.22 A few examples of sRNA-based regulation are described below.
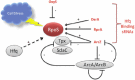
As a master regulator of stress response in E. coli the sigma factor RpoS (ss) responds to multiple stresses with almost 500 genes directly or indirectly under its control.17 In an example of RNA-based regulation (Fig. 2), RpoS is regulated by at least four small RNAs and requires the RNA chaperone protein Hfq.17 Two Hfq-binding sRNAs DsrA and RprA also positively regulate rpoS translation by base pairing to the upstream part of an inhibitory stem-loop in the rpoS mRNA leader sequence and freeing the ribosome binding site.17 OxyS, an sRNA expressed under oxidative stress, represses expression by a less understood mechanism (reviewed in ref. 17). In a manner similar to that of miRNA-target feedback loops seen in eukaryotes, one small RNA ArcZ directly represses and is repressed by arcB transcription, and is also a positive regulator of RpoS.17 It is suggested that each of these three positive regulators of RpoS (ArcZ, DsrA and RprA) have different strengths in their ability to open the RpoS structure, and thus may regulate RpoS to different levels.17 ArcZ has other targets than RpoS including tpx which encodes a lipid hydroperoxide peroxidise, and sdaC, a putative serine transporter in Salmonella.23 The complexity of this small RNA network is further increased if we add the negative regulation of tpx and arcB by ArcA, and that the ArcA/ArcB system is one of the central regulators in E. coli.17,24
The Hfq protein, as well as mediating small RNA interactions with their targets also has connections to RNA degradation via a multi-enzyme complex called the degradosome.25-27 The degradosome is made up primarily of four enzymes including the endoribnuclease RNase E, and the exoribonuclease PNPase.27 The degradosome appears widespread in bacteria, but a similar complex, the exosome is found in archaea and eukaryotes assembling on a core that resembles PNPase.25,28 Hfq can bind to the degradosome to promote RNase E cleavage and subsequent degradation of target mRNAs after base-pairing with small RNAs.27,29 An interesting feature to emerge is that the interaction between Hfq and RNase E is not direct, but is likely to be mediated by another, yet to be identified RNA.27 Hence, it is possible that the degradation of RNA may be regulated by other RNA species.
In another example of RNA-based networking, catabolite repression prevents transcription of genes required for a less desired substrate when more than one carbon source is available for a bacterial species. Monod30 first observed that E. coli and B. subtilis, when given two carbon sources at the same time, will often degrade one substrate first (the one giving faster growth) before the other (summarised in refs. 31, 32). In Pseudomonas aeruginosa, the small RNA CrcZ binds to the Crc protein, to come under the control of the CbrA/CbrB system.32 The CbrA/CbrB system adjusts CrcZ small RNA levels in response to different carbon sources allowing a gradual mode of catabolite repression operating at the post-transcriptional level.32 Another small RNA CyaR is regulated by the Crp gene (a global regulator of many sRNAs) to increase expression when cyclic AMP (cAMP) levels are high.14
sRNAs are heavily involved in the interaction of the bacterial cell with its environment. The outer membrane of the Gram-negative cell prevents toxic compounds from entering the cell,33 and contains channels formed from OMP porin proteins. Some of these OMP proteins are regulated by small RNAs, including OmpC regulated by the MicC RNA and OmpA regulated by MicA RNA.33 MicA requires Hfq both for its own intracellular stability and for annealing to the ompA mRNA.33 The OmpX protein (bacterial adhesion and mammalian cell interation protein) is down-regulated by the small RNA CyaR (discussed above) linking the starvation response to protein which aid in moving to a new source of nutrients.14
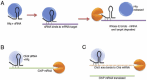
Another OMP-type protein ChiP (chitoporin) is down regulated by the small RNA ChiX.17,34 ChiX regulation of ChiP is different from the standard pathway as the sRNA does not become destablilised after base-pairing with its target (Fig. 3A) and it is thought that it could act catalytically to degrade multiple mRNAs (Fig. 3B).17 However, there is also competitive regulation of the ChiX sRNA by an mRNA decoy where base-pairing of ChiX to an intergenic region of the chb mRNA induces degradation of the sRNA (Fig. 3C). Upon induction by chitobiose, there is degradation of ChiX relieving the inhibition of the chip mRNA.17
sRNAs are important factors in the making and breaking of sugars in the cell.19 A classic example is the regulation of the ptsG gene which is part of the phosophotransferase system involved in the uptake of glucose. The ptsG gene transports and phosphoylates glucose to G6P and while G6P is essential for glucose metabolism, accumulation within the cell causes sugar-phosphate toxicity (also called phospho-sugar stress19). The SgrS gene codes for an Hfq-associated sRNA which has a dual function of regulating ptsG by base-pairing as well as coding for a small protein SgrT.35 SgrT has been shown to also block glucose uptake without affecting levels of ptsG mRNA or PtsG protein, although its exact mechanism is not yet known.35 It is interesting to note that the topological network properties associated with carbon metabolism are very different to those that control developmental processes, with short parallel transcriptional cascades for the former, and long intertwined regulatory cascades for the latter.18 Such a mechanism again allows for a rapid change in carbon source to be maximally exploited by the bacteria.
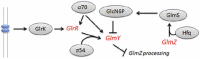
In another example where the mechanism is more known (Fig. 4), the synthesis of glucosamine-6-phosphate (GlcN6P) is regulated by two small RNAs GlmY and GlmZ36 (and reviewed in ref. 31). GlcN6p can be derived from amino sugars in the cell environment or by de novo synthesis with glucosamine-6-phosphate synthase (GlmS). GlmS is encoded with its partner GlmU in an operon and while GlmU is essential, GlmS is only required in the absence of amino sugars. The small RNA GlmZ is typically processed to be inactive. When GlcN6P is low, GlmY accumulates and inhibits processing of GlmZ. GlmZ then pairs with the glmS mRNA transcript activating GlmS which resynthesizes GlcN6P. The glmS mRNA is normally unstable due to secondary structure which buries its ribosomal binding site. However, base-pairing with sRNA GlmZ aided by the Hfq protein changes the structure and allows efficient translation of glmS. GlmY is also processed but how this affects the network is not yet characterised.36 In another interesting twist, GlmY is controlled by two overlapping promoters with the same transcription start site.36 One promoter activates transcription during the cell's exponential growth and the other operates during the cell's stationary phase. This may control the levels of GlcN6P from being high during the stationary phase when GlcN6P is not required (when cell wall and outer membrane synthesis is not required).36
Another interesting point about glmS is that the 5′-UTR contains a ribozyme (a catalytic RNA molecule that can cleave RNA), and functions as a (GlcN6P)-dependent catalyst enabling the riboswitch-like regulation of amino-sugars. In other words, the binding of GlcN6P has an effect on a gene involved in its production,37 but without the need for the riboswitch structural elements of an aptamer and expression platform. It has been shown that GlcN6P is absolutely required for the catalytic activity,38 demonstrating the property of RNA to be a metabolite-responsive ribozyme. Functionally, GlcN6P binding stimulates cleavage near the 5′ end of the transcript37 and furthermore the cleaved downstream product is targeted for intracellular degradation.39 RNase E is responsible for global mRNA decay in E. coli, but this enzyme is not present in a variety of prokaryotes e.g., B. subtilis. RNAse J1 has been shown fulfil the same role in B. subtilis, wherein it is responsible for the degradation of the 3′ cleavage product upon ribozyme action.39
These are only a few examples of identified small RNA based networks present within bacteria. With new sequencing techniques able to identify a genome's worth of RNAs in a single run, we expect there to be many others yet to be characterised. How these networks have evolved within prokaryotes is also another question wide open to be explored.
TRANSFER MESSENGER RNA
With new research the traditional roles of the main RNAs in the bacterial cell are being expanded. An example is tmRNA (transfer-messenger RNA also known as SsrA RNA or 10Sa RNA), a specialised tRNA molecule that along with a small protein (SmpB-small protein B), mediates the rescuing of stalled ribosomes. When incomplete or damaged mRNA is translated, the ribosome stalls leaving a peptidyl-tRNA on the P site of the ribosome. Alanine-charged tmRNA then enters the A-site of the stalled ribosome and accepts the peptide.40 The translation of the sequence encoded in the tmRNA resumes the translation process to add a C-terminal peptide tag to the protein then uses an in-frame stop codon to complete the translation.40-42 This process, known as trans-translation allows the tmRNA to act as both a tRNA and an mRNA as its name suggests.43 The secondary structure of the tmRNA holds the key to how it is able to serve both functions.41 One domain of the tmRNA has an amino acid acceptor stem chargeable with alanine and a T arm with modified nucleotides (as expected in a tRNA). However, the D arm of the tRNA does not fold-like a typical tRNA and there is no anticodon loop. Instead an mRNA-like domain (MLD) is located in a pseudoknot rich region and contains a short open reading frame with the tmRNA motif (AADENYALAA) followed by a typical stop codon. It is this domain that targets the truncated protein for degradation.41 Trans-translation appears to be conserved throughout bacteria and is also present in some mitochondria and chloroplasts.42,43 In some bacterial species the gene for the tmRNA (ssrA) is essential (e.g., Neisseria gonorrhoeae, Mycoplasma spp., Synechococcus spp.), but in other species it is merely important for surviving challenging growth conditions.41 As well as its typical tmRNA role tmRNA also binds in an antisense direction to the 5′ UTR of the crtMN mRNA, and thus may play an important role in the pathogenic nature of Staphylococcus aureus.44
tmRNAs are involved in sporulation in some bacterial species. Under starvation conditions B. subtilis enters sporulation where the cell undergoes asymmetrical division into a two chambered sporangium consisting of a forespore and a mother cell.40 The forespore matures, aided by the mother cell until the forespore is engulfed by the mother cell, forming a free protoplast surrounded by a double membrane inside the mother cell. During sporulation different genes are expressed in each compartment which is controlled by compartment specific RNA polymerase sigma factors σF and σG in the forespore and sE and sK in the mother cell (reviewed in ref. 40). tmRNA has been shown to be necessary for the later stages of sporulation, since only 5% of B. subtilis cells lacking the tmRNA gene produced heat resistant spores.40 As spore formation is regarded as a stress response to nutrient starvation, this process may also lead to increased ribosome stalling which is rescued by tmRNA. Without tmRNA, stalled ribosomes may accumulate resulting in the cell unable to translate essential spore-related proteins.40 This may also be the case for other stress responses since tmRNA deletion also leads to hypersensitity to drugs such as aza-C (5-azacytidine) which induces DNA-protein crosslinks between cytosine methyl transferase and DNA inhibiting methylation. This cross-linking is known to lead to blocking of the translation system, and tmRNA thus relieves the blockage.45
CONCLUSION
In this chapter we have described in some detail RNase P, small RNAs, sigma factors and transfer messenger RNAs and how they interact in networks within the prokaryotic cell. These networks are only a few coming to light from recent studies of ncRNAs in prokaryotes. Archaeal systems are yet to be fully explored and so are many species of bacteria that are distantly related to the model species (e.g., E. coli and B. subtilis). As these studies progress, in both model and non-model prokaryotes we may then be able to begin to unravel a clearer idea of the 'general' prokaryotic RNA-infrastructure. However, it is possible that there is such diversity amongst the bacterial and archaeal groups that such a general picture cannot be drawn. RNA studies are still in their early days in prokaryotes and there is no doubt that there is exciting work ahead.
ACKNOWLEDGEMENTS
LJC would like to thank Prof. David Penny for continued support, financial and intellectual and stimulating discussions around this topic. PJB would like to thank Prof. Peter Lockhart for his support, and is part funded by the Royal Society of New Zealand Marsden Fund (08-MAU-099).
REFERENCES
- 1.
- Esakova O, Krasilnikov AS. Of proteins and RNA: the RNase P/MRP family. Rna. 2010;16(9):1725–1747. [PMC free article: PMC2924533] [PubMed: 20627997]
- 2.
- Lai LB, Vioque A, Kirsebom LA, et al. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett. 2010;584(2):287–296. [PMC free article: PMC2799185] [PubMed: 19931535]
- 3.
- Marvin MC, Engelke DR. Broadening the mission of an RNA enzyme. J Cell Biochem. 2009;108(6):1244–1251. [PMC free article: PMC2800852] [PubMed: 19844921]
- 4.
- Marvin MC, Engelke DR. RNase P: increased versatility through protein complexity? RNA Biol. 2009;6(1):40–42. [PMC free article: PMC2798119] [PubMed: 19106627]
- 5.
- Cho IM, Lai LB, Susanti D, et al. Ribosomal protein L7Ae is a subunit of archaeal RNase P. Proc Natl Acad Sci U S A. 2010;107(33):14573–14578. [PMC free article: PMC2930468] [PubMed: 20675586]
- 6.
- Jarrous N, Gopalan V. Nucleic Acids Res. 2010. Archaeal/Eukaryal RNase P: subunits, functions and RNA diversification. [PMC free article: PMC3001073] [PubMed: 20716516]
- 7.
- Pulukkunat DK, Gopalan V. Studies on Methanocaldococcus jannaschii RNase P reveal insights into the roles of RNA and protein cofactors in RNase P catalysis. Nucleic Acids Res. 2008;36(12):4172–4180. [PMC free article: PMC2475606] [PubMed: 18558617]
- 8.
- Koutmou KS, Zahler NH, Kurz JC, et al. Protein-precursor tRNA contact leads to sequence-specific recognition of 5 leaders by bacterial ribonuclease P. J Mol Biol. 2010;396(1):195–208. [PMC free article: PMC2829246] [PubMed: 19932118]
- 9.
- Mohanty BK, Kushner SR. Ribonuclease P processes polycistronic tRNA transcripts in Escherichia coli independent of ribonuclease E. Nucleic Acids Res. 2007;35(22):7614–7625. [PMC free article: PMC2190699] [PubMed: 17981836]
- 10.
- Mans RM, Guerrier-Takada C, Altman S, et al. Interaction of RNase P from Escherichia coli with pseudoknotted structures in viral RNAs. Nucleic Acids Res. 1990;18(12):3479–3487. [PMC free article: PMC331000] [PubMed: 2194161]
- 11.
- Bai Y, Rider PJ, Liu F. Catalytic M1GS RNA as an antiviral agent in animals. Methods Mol Biol. 2010;629:339–353. [PubMed: 20387160]
- 12.
- Seif E, Altman S. RNase P cleaves the adenine riboswitch and stabilizes pbuE mRNA in Bacillus subtilis. Rna. 2008;14(6):1237–1243. [PMC free article: PMC2390808] [PubMed: 18441052]
- 13.
- Altman S, Wesolowski D, Guerrier-Takada C, et al. RNase P cleaves transient structures in some riboswitches. Proc Natl Acad Sci U S A. 2005;102(32):11284–11289. [PMC free article: PMC1183601] [PubMed: 16061811]
- 14.
- De Lay N, Gottesman S. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J Bacteriol. 2009;191(2):461–476. [PMC free article: PMC2620814] [PubMed: 18978044]
- 15.
- Gottesman S, Storz G. Cold Spring Harb Perspect Biol. 2010. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. [PMC free article: PMC3225950] [PubMed: 20980440]
- 16.
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136(4):615–628. [PMC free article: PMC3132550] [PubMed: 19239884]
- 17.
- Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. Embo J. 2010;29(18):3094–3107. [PMC free article: PMC2944060] [PubMed: 20683441]
- 18.
- Martinez-Antonio A, Janga SC, Thieffry D. Functional organisation of Escherichia coli transcriptional regulatory network. J Mol Biol. 2008;381(1):238–247. [PMC free article: PMC2726282] [PubMed: 18599074]
- 19.
- Gorke B, Vogel J. Noncoding RNA control of the making and breaking of sugars. Genes & development. 2008;22(21):2914–2925. [PubMed: 18981470]
- 20.
- Masse E, Salvail H, Desnoyers G, et al. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10(2):140–145. [PubMed: 17383226]
- 21.
- Svenningsen SL, Tu KC, Bassler BL. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J. 2009;28(4):429–439. [PMC free article: PMC2632942] [PubMed: 19165149]
- 22.
- Shimoni Y, Friedlander G, Hetzroni G, et al. Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol Syst Biol. 2007;3:138. [PMC free article: PMC2013925] [PubMed: 17893699]
- 23.
- Papenfort K, Said N, Welsink T, et al. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol. 2009;74(1):139–158. [PubMed: 19732340]
- 24.
- Soper T, Mandin P, Majdalani N, et al. Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci U S A. 2010;107(21):9602–9607. [PMC free article: PMC2906882] [PubMed: 20457943]
- 25.
- Hardwick SW, Chan VS, Broadhurst RW, et al. Nucleic Acids Res. 2010. An RNA degradosome assembly in Caulobacter crescentus. [PMC free article: PMC3045602] [PubMed: 20952404]
- 26.
- Janga SC, Babu MM. Transcript stability in the protein interaction network of Escherichia coli. Mol Biosyst. 2009;5(2):154–162. [PubMed: 19156261]
- 27.
- Worrall JA, Gorna M, Crump NT, et al. Reconstitution and analysis of the multienzyme Escherichia coli RNA degradosome. J Mol Biol. 2008;382(4):870–883. [PMC free article: PMC7611026] [PubMed: 18691600]
- 28.
- Evguenieva-Hackenberg E, Klug G. RNA degradation in Archaea and Gram-negative bacteria different from Escherichia coli. Prog Mol Biol Transl Sci. 2009;85:275–317. [PubMed: 19215775]
- 29.
- Kaberdin VR, Lin-Chao S. Unraveling new roles for minor components of the E.coli RNA degradosome. RNA Biol. 2009;6(4):402–405. [PubMed: 19667755]
- 30.
- Monod J. Paris: Hermann & cie. 1942. Recherches sur la croissance des cultures bacteriennes.
- 31.
- Fujita Y. Carbon catabolite control of the metabolic network in Bacillus subtilis. Biosci Biotechnol Biochem. 2009;73(2):245–259. [PubMed: 19202299]
- 32.
- Sonnleitner E, Abdou L, Haas D. Small RNA as global regulator of carbon catabolite repression in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2009;106(51):21866–21871. [PMC free article: PMC2799872] [PubMed: 20080802]
- 33.
- Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol. 2006;9(6):605–611. [PubMed: 17055775]
- 34.
- Figueroa-Bossi N, Valentini M, Malleret L, et al. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes & development. 2009;23(17):2004–2015. [PMC free article: PMC2751969] [PubMed: 19638370]
- 35.
- Wadler CS, Vanderpool CK. Characterization of homologs of the small RNA SgrS reveals diversity in function. Nucleic Acids Res. 2009;37(16):5477–5485. [PMC free article: PMC2760806] [PubMed: 19620214]
- 36.
- Reichenbach B, Gopel Y, Gorke B. Dual control by perfectly overlapping sigma 54- and sigma 70- promoters adjusts small RNA GlmY expression to different environmental signals. Mol Microbiol. 2009;74(5):1054–1070. [PubMed: 19843219]
- 37.
- Winkler WC, Nahvi A, Roth A, et al. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428(6980):281–286. [PubMed: 15029187]
- 38.
- McCarthy TJ, Plog MA, Floy SA, et al. Ligand requirements for glmS ribozyme self-cleavage. Chem Biol. 2005;12(11):1221–1226. [PubMed: 16298301]
- 39.
- Collins JA, Irnov I, Baker S, et al. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007;21(24):3356–3368. [PMC free article: PMC2113035] [PubMed: 18079181]
- 40.
- Abe T, Sakaki K, Fujihara A, et al. tmRNA-dependent trans-translation is required for sporulation in Bacillus subtilis. Mol Microbiol. 2008;69(6):1491–1498. [PubMed: 18673456]
- 41.
- Wower J, Wower IK, Zwieb C. Making the jump: new insights into the mechanism of trans-translation. J Biol. 2008;7(5):17. [PMC free article: PMC2447533] [PubMed: 18598387]
- 42.
- Une M, Kurita D, Muto A, et al. Trans-translation by tmRNA and SmpB. Nucleic Acids Symp Ser (Oxf). 2009;(53):305–306. [PubMed: 19749382]
- 43.
- Hayes CS, Keiler KC. Beyond ribosome rescue: tmRNA and co-translational processes. FEBS Lett. 2010;584(2):413–419. [PMC free article: PMC2794900] [PubMed: 19914241]
- 44.
- Liu Y, Wu N, Dong J, et al. FEBS Lett. 2010. SsrA (tmRNA) acts as an antisense RNA to regulate Staphylococcus aureus pigment synthesis by base pairing with crtMN mRNA. [PubMed: 20854817]
- 45.
- Kuo HK, Krasich R, Bhagwat AS, et al. Importance of the tmRNA system for cell survival when transcription is blocked by DNA-protein cross-links. Mol Microbiol. 2010;78(3):686–700. [PMC free article: PMC2963719] [PubMed: 20807197]
- RNA NETWORKS IN PROKARYOTES II: tRNA Processing and Small RNAs - Madame Curie Bi...RNA NETWORKS IN PROKARYOTES II: tRNA Processing and Small RNAs - Madame Curie Bioscience Database
- DNA Packaging by Bacteriophage P22 - Madame Curie Bioscience DatabaseDNA Packaging by Bacteriophage P22 - Madame Curie Bioscience Database
- Regulation of Insulin Action and Insulin Secretion by SNARE-Mediated Vesicle Exo...Regulation of Insulin Action and Insulin Secretion by SNARE-Mediated Vesicle Exocytosis - Madame Curie Bioscience Database
- Structure, Function and Assembly of Flagellar Axial Proteins - Madame Curie Bios...Structure, Function and Assembly of Flagellar Axial Proteins - Madame Curie Bioscience Database
- The ø29 DNA Packaging Motor: Seeking the Mechanism - Madame Curie Bioscience Dat...The ø29 DNA Packaging Motor: Seeking the Mechanism - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...