NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
The Tn4371 ICE (Integrative Conjugative Element) family refers to a group of mobile genetic elements with four modules containing genes involved in integration (via a tyrosine-based site-specific recombinase), maintenance/stability, accessory genes conferring a special phenotype to the host bacteria and genes involved in conjugational transfer. The latter display similarity with conjugative genes of Ti plasmids and IncP broad host range plasmids. Currently this ICETn4371 family harbours around 40 elements with sizes ranging from 38 to 101 kb. Elements carrying accessory genes, which are mostly flanked by the conjugative genes rlxS (virD2) and traR, reside in Beta- and Gammaproteobacteria, while elements in Alphaproteobacteria apparently lack accessory genes and display a more divergent pattern. Accessory genes have very diverse functions including catabolism of xenobiotic compounds, resistance to heavy metals, to antibiotics, chemolithotrophic metabolism, but also some unknown functions. Strains from man-made environments (sewages, industrial wastes and clinical settings) are predominant among the bearers of Tn4371 ICEs next to plant pathogens, which are also well represented. Insights into ~30 characteristic genes (such as those encoding site-specific recombinases and excisionases) as well as their distribution in the described elements and their relationship with other mobile genetic elements is provided.
Introduction
Tn4371 is a 55 kb mobile element, which allows its host to degrade biphenyl and 4-chlorobiphenyl. It was originally isolated after mating between Cupriavidus oxalaticus A5 (formerly Alcaligenes eutrophus A5) carrying the broad host range conjugative plasmid RP4 and Cupriavidus metallidurans (formerly Ralstonia metallidurans) CH34. Transconjugants were selected by their resistance to heavy metals, which is a key trait of CH34, and their ability to grow on biphenyl as a sole source of carbon and energy.1 These transconjugants carried an RP4 plasmid with a 55 kb insert near its tetracycline resistance operon. The element was designated Tn4371 as it was shown to transpose to other locations,1,2 and subsequently sequenced and studied further.3 Closely related elements have been found in the genome sequences of a number of bacteria in the Beta- and Gammaproteobacteria classes including in Cupriavidus metallidurans CH34 and Pseudomonas aeruginosa.3-6 Table 1 shows a full list of all elements.
Table 1
ICE Tn4371 elements identified in different Proteobacteria.
Integrative Conjugative Elements (ICEs) carry functional modules involved in their conjugative transfer, chromosomal integration and for control of expression of ICE genes. They establish themselves in their host genome at a unique or limited number of sites via site-specific integration. First discovered in the genomes of various Firmicutes, ICEs have been found in various Alpha, Beta- and Gammaproteobacteria, and Bacteroides species.7 The first ICE found, Tn916 from Streptococcus faecalis8 (now Enterococcus faecalis), has a broad transfer range.9,10
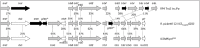
As with other ICEs, those in the Tn4371 family are characterized by a mosaic structure. They consist of four modules, three conserved ones, including an integration/excision module (IntMod), a stabilization/maintenance module (StaMod) and a Ti-RP4-like transfer module (TraMod), and a variable module with accessory genes (AccMod) (Fig. 1).4,5 Many of the elements have been found integrated into a conserved host attB site with the 5′-TTTTTYAT-3′ sequence.2 The ends of the elements can be detected covalently linked as a transfer intermediate indicating that Tn4371 transposition most likely involves a site-specific integration/excision process.2,4 Their size varies from 38 to 101 kb depending on the number of accessory genes present. All together, the conserved genes from IntMod, StaMod and TraMod account for approximately 24 kb (a 1.5 kb IntMod, a 8.5 kb StaMod and a 14 kb TraMod).
Taxonomy of Tn4371-Like ICEs and Their Hosts
Elements that carry genes characteristic to Tn4371 (see Fig. 2) and a main AccMod between rlxS and traR (see below), have been grouped as ICETn4371-elements. This large group also includes elements with more divergent tyrosine-based site-specific recombinases (TBSSRs) previously identified by Ryan et al.4 The nomenclature system designed by Ryan et al.,4 which was based on an adaptation of the system used for naming transposons described by Roberts et al.,11 was preserved and continued.
Previous in silico comparative studies identified ICEs closely related to Tn4371 in the genomes of several Betaproteobacteria. Three elements were identified in C. metallidurans CH34,3,5 two in Delftia acidovorans SPH-14,5 and Bordetella petrii DSM12804,4,5,12 and a single one in Acidovorax avenae subsp citrulli AAC00-1, Acidovorax ebreus TPSY, Acidovorax sp JS42, Burkholderia pseudomallei MSHR346, Comamonas testosteroni KF-1, Polaromonas naphthalenivorans CJ2 plasmid pPNAP01, Ralstonia pickettii 12J and Ralstonia solanacearum GMI1000.3,4 Tn4371-like elements were also found in Gammaproteobacteria including Azotobacter vinelandii DJ, Congregibacter litoralis KT71, Dickeya dadantii 3937 (formerly Erwinia chrysanthemi3), P. aeruginosa strains 2192, PA7, PACS171b and UCBPP-PA14, Shewanella sp ANA-3, Stenotrophomonas maltophilia K279a and Thioalkalivibrio sp HL-EbGR7.3,4
This chapter also includes (or further characterizes) new elements in the Betaproteobacteria such as Acidovorax sp KKS102, Alicycliphilus denitrificans BC, Burkholderia ambifaria AMMD, Burkholderia multivorans strains CGD2, CGD2M and ATCC 17616, and R. solanacearum MolK2. The Gammaproteobacteria included Citrobacter sp 30_2, Pectobacterium carotovorum subsp brasiliensis PBR1692, P. aeruginosa 39016 and Xanthomonas fuscans subsp aurantifolii str. 11122 (Table 1). New elements were also discovered in the Alphaproteobacteria and will be discussed below.
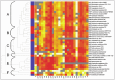
When a set of characteristic proteins from the IntMod, StaMod and TraMod of the ICETn4371 elements was collected and used to perform pairwise alignments and calculate protein similarities with the original Tn4371 (Fig. 2) ParA and TraG emerged as the most conserved proteins followed by proteins of the Trb cluster.
Clustering analysis of all these pairwise alignments produced 3 main groups. A large cluster with multiple subgroups (A, B, C and D) contained most of the elements and within this large cluster, subgroup A (Fig. 2) contained elements from Beta- and Gammaproteobacteria, which belong to a variety of environments, strongly suggesting that ICETn4371 elements may have a broad range of transfer and replication abilities. Subgroup B contained four from Gamma- and two from Betaproteobacteria, while subgroup C contained seven elements from Betaproteobacteria and one from Gammaproteobacteria. All these strains originate from environmentally polluted or industrial settings.
A second large cluster (group E in Fig. 2) included ICETn43716037 from D. acidovorans SPH-1, ICETn43716038 from C. testosteroni KF-1, ICETn43716033 from R. pickettii 12J, ICETn43716039 from Acidovorax sp JS42, ICETn43716064 from B. pseudomallei MSHR346, and ICETn43716055 and ΔICETn43716056 from C. metallidurans CH34, for which the corresponding hosts are Betaproteobacteria isolated from waste waters or again from industrial environments.
The last cluster (group F in Fig. 2) grouped the elements from Xanthobacter autotrophicus Py2 (ICETn43716139), D. dadantii 3937 (ΔICETn43716154) and B. petrii DSM12804 (ICETn43716141) and appeared to be more distant from all the other groups. The element from X. autotrophicus Py2 is shown here as a representative of ICETn4371-like elements that we have observed in a large group of Alphaproteobacteria. These elements do not contain an AccMod between rlxS and traG nor a traR gene, have a shorter rlxS gene, and contain a slt gene coding for a lytic transglycosylase. The latter is so far specific for ICETn4371-like elements in Alphaproteobacteria and located between traF and rlxS. The large group of related ICETn4371 variants in Alphaproteobacteria is interesting as genes in the StaMod and the TraMod of the ICEMlSymR7A element from the Alphaproteobacterium Mesorhizobium loti R7A are also highly similar to ICETn4371 (see below) indicating that there may be a wide range of relationships with at least one other family of elements. Other Alphaproteobacteria that carry ICETn4371-like elements similar to X. autotrophicus Py2 (ICETn43716139) include Oligotropha carboxidovorans OM5, Rhodopseudomonas palustris DX1, Gluconacetobacter diazotrophicus PAl5, Chelativorans sp BNC1 (formerly Mesorhizobium), Azorhizobium caulinodans ORS571, Bradyrhizobium japonicum USDA110, Bradyrhizobium sp BTAi1, Caulobacter sp K31 and Parvibaculum lavamentivorans DS1 among others. Very likely, further studies will show that cluster F may further subdivide in various subgroups.
In various strains that contain two or more ICETn4371 elements, the corresponding elements belong to different groups: C and E for D. acidovorans SPH-1, Acidovorax sp JS42 and C. metallidurans CH34, A and C for A. denitrificans BC, D and F for B. petrii DSM12804. This break down of elements into groups could be an example of a phenomenon known as entry exclusion that is seen in some families of ICEs most notably the R391/SXT family found in Enterobacteriaceae.13 These ICEs can be divided into two exclusion groups, called S and R.14 Bacteria that host an S group ICE are less susceptible to acquire another S group ICE while the acquisition of an R group ICE is not restricted. This exclusion activity is controlled by the TraG protein and a protein termed the entry exclusion protein, Eex (EexR or EexS based on the exclusion group).15 These are inner-membrane proteins expressed in donor and recipient cells, respectively.16 An entry exclusion system is also found in IncPα plasmid RP4 (to which the transfer system of ICETn4371 is related). The trbK gene in the Tra2 region of the plasmid has been shown to code for the single protein involved in entry exclusion. This small hydrophilic polypeptide of 69 amino acid residues contains a lipoprotein signature.17 Four lipoproteins (gilA1[genomic island lipoprotein A1], gilA2, gilB1 and gilB1) are found in the ICETn4371 elements (see below) but none of these proteins show homology to TrbK. Although Figure 2 shows that the GilA2 proteins of group E are highly identical with GilA2 of the original Tn4371 and as such quite distinct from the other groups, further investigation is required to determine if these groupings are an example of entry exclusion and if so what is the associated mechanism.
In an alternative approach, separate phylogenetic trees were constructed for each of the characteristic proteins using the neighbor-joining method. To explore the underlying phylogenomic information, these (partially) overlapping data sets were used to construct a supertree using Clann Software.18 However, bootstrap analysis did not support such a supertree (data not shown). Thus the individual trees generated from ICETn4371 signature proteins are not congruent, suggesting that different protein families have been subjected to different evolutionary processes.
Integration and Excision (IntMod)
The IntMod makes up the left part of the ICETn4371 and generally begins with an int gene coding for a tyrosine-based site-specific recombinase (TBSSR). In some elements the int gene is followed by a few accessory genes (observed in around 20% of the elements). These have an unknown function (ICETn43716055 and ICETn43716056) or are involved in heavy metal resistance as in D. acidovorans SPH1 ICETn43716067. These accessory genes will be further described in the section about accessory genes.
Tyrosine-based site-specific recombinases are well known DNA breaking-rejoining enzymes, which are major actors in the comings and goings of mobile genetic elements in bacterial genomes. The 3D structure and molecular mechanisms of action of several enzymes of the family are well documented.19-24 TBSSRs are usually called "integrases" since they catalyze integration and excision of phages, integron cassettes,25 ICEs13 and other genomic islands.24,26 However some, the XerCD enzymes, resolve chromosome dimers to ensure successful bacterial circular chromosome segregation, plasmid di/multimers or cointegrates generated by some transposons (for a review see ref. 27). TBSSRs specifically bind to their cognate att sites and usually need assistance from element-coded directionality proteins called excisionases (Xis) and host-coded nucleoid associated proteins such as IHF or HU for the catalysis of excision.
ICETn4371 Int Proteins
As shown in Figure 1, a TBSSR gene lies adjacent to the left end/attL site of ICETn4371 elements. Ryan et al.4 reported that ICETn4371 TBSSRs and att sites are well conserved and more related among them than they are to those encoded by phages and other ICEs. This is further supported by the grouping of most of the ICETn4371 TBSSR proteins into a single clique upon clustering of all bacterial proteins in the NCBI RefSeq repository (CLSK897396).28 However, multiple alignments of all TBSSRs encoded by the ICETn4371 described in Table 1 showed that seven sequences deviate from the very well conserved bulk, both overall and in the probable C-terminal catalytic motif (RHR-Y, aa 353–385, for the bulk; probably RRT-Y, aa 363–397, for five of the diverging ones) (Fig. 3). These seven proteins are from R. solanacearum MolK2 (ICETn43716143; YP_002253784), B. petrii DSM12804 (ICETn43716141; YP_001633162), C. metallidurans CH34 (ICETn43716055; YP_583617), A. ebreus TPSY (ICETn43716140; YP_002552729), B. ambifaria AMMD (ΔICETn43716151; YP_773911) and D. dadantii 3937 (ΔICETn43716154). The left end of the latter element carries two TBSSRs coding genes, one of which is more related to the other TBSSRs and hence appears as a more likely candidate for being the element's integrase. The variation in IntMod within an ICE family is not unique. Tn5397, a 21 kb ICE from Clostridium difficile, shares sequence similarity with Tn916 but bears a serine-based site-specific recombinase (TndX) instead of the TBSSR present in Tn916.29
The strong clustering (with almost 100% identity) of five TBSSRs found on elements with different AccMods but residing in hosts isolated from similar environments (wastes, industrial areas) suggests recent horizontal gene transfer and rearrangements. These features and the presence of divergent IntMods associated with a conserved StaMod and TraMod clearly points to the modular structure of ICETn4371.
ICETn4371 Xis Proteins: Excisionases
Phage excisionases are poorly conserved, short and basic,32 and only few of their homologs in other mobile genetic elements have been characterized (see for instance ref. 33, TorI). Ryan et al.4 suggested that ORF00035 of Tn4371 (CAD61129) and its orthologs on other ICETn4371 are excisionases because they show sequence similarity to the RdfS protein of ICEMlSymR7A, the symbiosis island from M. loti R7A.34 This RdfS protein (recombination directionality factor or excisionase) was shown to be necessary for efficient excision.34 In addition, ORF00034 of Tn4371 located upstream of and in the same orientation as rdfS codes for a protein similar to a fused product of msi172 and msi171 of ICEMlSymR7A, which are required for excision and transfer of ICEMlSymR7A.35 An ortholog of ORF00034 is found in all ICETn4371 discovered to date and has been called the excision associated protein (eap).35 Expression of msi172 and msi171 is regulated by quorum sensing via an N-acyl-L-homoserine lactone LuxR/LuxI (TraR/TraI) system, which is not present in ICETn4371 elements.
Ramsay et al.35 hypothesized that this regulation by TraR/TraI may be a recent addition to an older regulation of excision such that this event is now coupled with ICEMlSymR7A transfer in response to autoinducer signaling. Such a coupling between excision and transfer would be beneficial to ICE elements in general. In the ICETn4371 family there is yet no experimental evidence for such a coupling.
One possibility would be that the last gene before attR, which has been designated gir (genomic island right), is also required for excision. This conserved gene codes for a protein of unknown function and has occasionally been annotated as a transfer gene because it appears to belong to the putative trb operon. However, the Gir protein has about 80 residues and a calculated isoelectric point around eight, which are properties consistent with those expected for an excisionase or a nucleoid associated protein.
The Attachment Site (att)
When the Tn4371 DNA sequence is compared with complete bacterial genomic sequences, the left end comes out very similar in all genomes containing the ICETn4371 elements described in Table 1 except six, which are those that have different TBSSRs. While most alignments cover at least 1347 bp from the very first Tn4371 nucleotide at the left end (i.e., the left end and most of the int gene, bases 181..1389), five start somewhat further (though still before the int start codon), consistent with the slightly more distant position of their cognate enzyme in the tree of the ICETn4371 TBSSRs. For the six sequences that have the most diverging integrases, there is no significant automatic alignment with Tn4371 left end. In addition when the putative C. metallidurans CH34 ICETn43716055 left end and int gene (bp 1,585,250–1,586,550) are used as query for a Blastn comparative search, only one among the five remaining elements (A. ebreus TPSY ICETn43716140; bp 1,324,867–1,326,166) shows up within the best hits.
Upon integration, the attTn site is normally split between the left and right ends of the integrated ICEs. Nucleotide alignments between Tn4371 right end and other ICEs in Table 1 are less straightforward. They usually cover the entire trb region and only four ICETn4371 align up to the end of the Tn4371 right end (nucleotide 54657).
Many of the elements have been found to integrate into a conserved host attB site with the 5′-TTTTTYAT-3′ sequence,2 however, the deviating 5′-TTTTTYGT-3′ sequence has been occasionally observed like for ICETn43716042, ICETn43716054 and ICETn43716068.
Transcriptional Units
At least ten int genes (from ICETn43716033, 6036, 6037, 6038, 6054, 6056, 6066, 6069, 6071 and 6142) appear to form a two-gene transcriptional unit (4bp overlap) with a conserved gene coding for a secreted protein of unknown function. A similar coupling (TBSSR coupled to a putative repressor gene) has already been described in plasmid pMOL28 and pMOL30 from C. metallidurans CH34 in which these units flank a genomic island involved in chromate resistance and a czc-like cluster involved in the resistance to Cd2+, Zn2+ and Co2+, respectively. The latter is fully conserved in R. pickettii 12J.5,6
Stability and Maintenance Genes (StaMod)
Downstream from the IntMod, a gene coding for the GilA1 lipoprotein and the genes yafZ and parB are present in most elements. YafZ (often annotated as F-plasmid gene 32-like protein) and ParB are related to proteins encoded by genes located near the transfer origin of Escherichia coli F plasmid. The function of YafZ is unknown, while ParB is similar to ParB-like nucleases initially identified as crucial for the proper partitioning of plasmid DNA during cell division in the absence of selection pressure.36-38 Additional plasmid-related genes are located downstream of a cluster containing gilA2 (coding for a lipoprotein and similar to gilA1) and the genes eap and rdfS involved in excision. These plasmid-related genes code for a RepA (replication) protein similar to various plasmid encoded replication proteins,3 the ParA partition protein of the type Ib family39 and an associated ParG protein. Rep and Par proteins have been proposed to act as a stabilization system for the maintenance of mobile elements in bacterial genomes,38,40 similar to the toxin-anti-toxin system (mosAT) coded by ORFs s052 and s053 of the SXT-ICE.41 P. aeruginosa ICE PAPI-1 contains an ortholog of the plasmid and chromosome partitioning system soj (parA) and deletion of soj from PAPI-1 resulted in the complete loss of the PAPI-1 pathogenicity island. The mechanism by which the Soj protein promotes PAPI-1 maintenance remains to be elucidated.42 Genes similar to soj have also been found in ICEHin1056 (Hemophilus influenzae),43 ICEA (Mycoplasma agalactiae strain 5632)44 and (although with a low identity) in the major genomic island CMGI-1 of C. metallidurans CH34. The parA and parG genes are part of a four-gene operon with 4 bp overlaps (suggesting transcriptional coupling) that also includes a gene of unknown function (gicC for genomic island conserved; see also below) and, rather unexpectedly, traF involved in conjugational transfer. Syntenic annotation (via the MaGe45 annotation platform from Genoscope) shows that this structure is maintained in almost all ICETn4371 including those of the Alphaproteobacteria. This four-gene operon will be further discussed in the TraMod section as well as the rlxS (virD2) gene coding for the relaxase protein that is essential in the conjugation process.
A rather large region in the middle of the StaMod between parB and gilA2 appears to be very variable from element to element with a variety of genes coding for small hypothetical proteins. Nevertheless, as shown in Figure 1 three small genes are apparently conserved in this region, namely gicB (RO00015), xre (RO00055) and gicA (RO00018).3 The gicB gene is located immediately downstream of parB and codes for a protein that is conserved in a large part of the ICEs reported in Table 1. The xre gene codes for a well recognizable transcriptional regulator.4 The gicA gene is remarkably conserved with very high identity in all the ICETn4371 including the related ICEs found in Alphaproteobacteria. The ubiquity and high sequence conservation of this ortholog inside this family suggests it codes for an essential function in ICETn4371 and ICETn4371-like elements.
The Accessory Genes (AccMod)
The great majority of accessory genes of the ICETn4371 elements are carried by the diverse AccMods but, more rarely, some are also associated with IntMods (and exceptionally to a StaMod like a gene coding for a cation diffusion factor in D. acidovorans SPH1). Accessory genes associated with IntMods include putative DNA helicases and nucleases, proteins with β-lactamase domains, putative reductases, insertion sequences, putative ubiquitin-activating enzymes, putative transcriptional regulators and many different hypothetical proteins whose functions are unknown. More defined systems are discussed below. The main AccMods are systematically located between two ORFs coding for the relaxase protein RlxS and a transcriptional regulator TraR, respectively. The traF gene, which codes for a protease, always lies upstream of the rlxS gene. The traR gene is followed by traG (virD4) coding for the conjugative coupling protein. The sequence between the rlxS gene and the first gene of the variable region, in all elements, is similar to the sequence of a region of Tn5 [U00004, 3787-4143] between the gene for streptomycin phosphotransferase and the transposase. This indicates that the diversity in this region of the element may be due to one or a number of Tn5-mediated insertion events.
The accessory genes vary from element to element and code for various traits such as degradative or metabolic properties, resistance mechanisms to either antibiotics or heavy metals, all of which are advantageous under certain environmental conditions. Yet some functions are hard to define while other AccMods are clearly a mosaic of different functions. Some gene groups are flanked by IS or/and carried by transposons. The main AccMod categories are often clearly recognizable and will be described below.
ICETn4371 Elements Coding for Resistance to Antibiotics
ICETn43716042 from P. aeruginosa PA7 (a clinical wound isolate) carries three genes involved in multiple antibiotic resistance genes (aminoglycoside 3′-phosphotransferase, streptomycin 3′-phosphotransferase, bleomycin resistance). Major Facilitator Superfamily (MFS) transporters and putative determinants for resistance to antibiotics are present in ICETn43716144 from A. vinelandii DJ (chloramphenicol and b-lactam) and ICETn43716075 from Shewanella sp ANA-3 (chloramphenicol). ICETn43716035 from C. litoralis KT71 carries a Resistance-Nodulation-Cell Division (RND) transporter (subfamily HAE5-RND) related to those involved in acriflavine resistance. The three component RND transporters are made of a cytoplasmic membrane export system, a membrane fusion protein, and an outer membrane factor and extrude substrates via an H+ antiport mechanism.46 The RND transporter of ICETn43716041 from P. aeruginosa 2192 is almost identical (99% protein identity) to that of ICETn43716035 from C. litoralis KT71. ICETn43716069 from S. maltophilia K279a carries a putative MFS transporter related to the emrAB system from E. coli,47 which usually functions as a specific exporter for certain classes of antimicrobial agents.
ICETn4371 Elements Coding Various Other Transporters
Next to the genes involved in antibiotic resistance, ICETn43716042 from P. aeruginosa PA7 carries a group of six genes coding for a KDP ATP-driven potassium transport system in which the KdpFABC complex acts as a high affinity K+ uptake system. In E. coli, the complex is synthesized when the constitutively expressed low affinity K+ uptake systems Trk and Kup no longer meet the cell's demand for potassium when external K+ becomes limiting.48,49 ICETn43716071 from Thioalkalivibrio sp HL-EbGR7 also contains a putative kdpFABC system, which is very similar (88% aa identity) to that of ICETn43716042 from P. aeruginosa PA7. ICETn43716039 from Acidovorax sp JS42 contains genes coding for a multidrug efflux pump, which appears inactivated by one of the many insertion sequences carried by this element. The partial element ΔICETn43716151 element from Burkholderia amfibaria AMMD codes for an ATP-binding cassette (ABC) transporter-like protein and a MFS transporter. The partial element ΔICETn43716153 in Citrobacter sp 30_2 contains multiple MFS transporters, a RND transporter (HAE3-RND family), a Cation Diffusion Facilitator (CDF) transporter and a P-type ATPase. ICETn43716145 of P. carotovorum subsp brasiliensis PBR1692 contains genes coding for a MFS transporter and an excinuclease ABC. The accessory genes of ICETn43716147 from Xanthomonas fuscans subsp aurantifolii str. ICPB 11122 code for multiple MFS transporters. ICETn43716146 from P. aeruginosa 39016 contains a RND transporter (uncharacterized family) and an ABC-type transporter. ICETn43716070 from P. aeruginosa PACS171b contains genes coding for a sulfate permease and a universal stress protein UspA. These genes share respectively 69 and 90% protein identity with those on transposon Tn6050 from C. metallidurans CH34.5
ICETn4371 Elements Encoding for Metal Resistance
ICETn43716038 from C. testosteroni KF-1 and ICETn43716037 from D. acidovorans SPH-1 are almost identical (98% DNA similarity) and accessory genes code for a Czc-like RND transporter, which probably belongs to the HME3b- or HME4-RND family.50 The second ICETn4371 element in D. acidovorans SPH-1 (ICETn43716067) carries genes involved in copper resistance (copSR copAB copGOF copCD copK) similar to the partial element ΔICETn43716152 from B. multivorans ATCC 17616 except the putative isoprenyl cysteine carboxyl methyltransferase. In addition, multiple heavy metal resistance clusters are located in the IntMod including resistance to silver/copper (HME4-RND family transporter), lead/cadmium (P-type ATPase and phosphatase), mercury (reductase) and arsenic (reductase and efflux). A CDF family transporter in the StaMod completes this diversified arsenal of metal resistance genes carried by ICETn43716067, which is probably the best example of an ICETn4371 specialized in the multiple resistances to heavy metals. These various gene clusters in ICETn43716067 contain the basic resistance genes without the diversity of satellite genes that is found in C. metallidurans CH34. ICETn43716148 from Alicycliphilus denitrificans BC contains a heavy metal P-type ATPase family and a czc-like RND transporter (family HME1-RND). ICETn43716141 from B. petrii DSM12804 contains multiple heavy metal resistance mechanisms including a silP-like gene coding for a P-type ATPase and a RND transporter putatively involved in Ag+/Cu+ resistance (family HME4-RND), a cluster involved in copper resistance (copDCBARS) and a czc-like RND transporter (family unclassified). In addition, the element carries accessory genes coding for arsenic resistance in the IntMod near the site-specific recombinase, which is also observed for ICETn43716057, ICETn43716067, ICETn43716070 and ΔICETn43716152.
ICETn43716066 from A. ebreus TPSY contains two magnesium transporters (MgtC and P-type ATPase family) and a czc-like RND transporter (family HME1-RND). ICETn43716069 from P. aeruginosa UCBPP-PA14 contains a czc-like RND transporter (family HME1-RND).
ICETn4371 Elements Coding for Catabolic Gene Products
ICETn43716065 (on plasmid pPNAP01 from P. naphthalenivorans CJ2) and ICETn4371KKS of Acidovorax sp KKS102 contain genes involved in biphenyl degradation similar to those found in the original Tn4371.4 However, for ICETn43716065 these genes are located in the IntMod while the AccMod contains genes involved in lipooligosaccharide metabolism, which could putatively have a role in tolerance toward desiccation,51 and multidrug resistance (family HAE1-RND). ICETn43716057 from B. multivorans CGD2, which is 100% identical to that of B. multivorans CGD2M, ICETn43716064 from B. pseudomallei MSHR346 and ICETn43716069 from S. maltophilia K279a are putatively involved in the degradation of aromatic compounds.4 ICETn43716033 from R. pickettii 12J and ICETn43716040 from B. petrii DSM12804 code for proteins putatively involved in lipid metabolism and threonine degradation, respectively. C. metallidurans CH34 carries three ICETn4371 elements with different accessory genes. ICETn43716054 carries genes involved in the degradation of aromatic compounds and a second cluster involved in hydrogenotrophy flanked by two copies of the same IS element (ISRme5). The latter putatively mediated the integration of this cluster into ICETn43716054.52 A second element (ICETn43716055) contains genes involved in hydrogenotrophy and fixation of carbon dioxide through the Calvin-Benson-Bassham cycle.5 The complete AccMod is flanked by two copies of IS1071 and loss of autotrophic growth occurs through IS1071-mediated excision.52
ICETn4371 Elements Coding for Multiple Functions and with Mosaic Structure
Numerous ICETn4371 elements described above carry genes that code for multiple functions such as ICETn43716042 (antibiotic resistance and K+ transport), ICETn43716054 (hydrogenotrophy and aromatic compound degradation), ICETn43716055 (hydrogenotrophy and carbon dioxide fixation), ICETn43716065 (multidrug resistance, biphenyl degradation and lipooligosaccharide metabolism) and ICETn43716069 (transporter, aromatic compound degradation). ICETn43716036 from A. avenae subsp citrulli AAC001 contains multiple IS elements, a MFS transporter and metabolic genes with unknown function. All genes except one just upstream traR, which encodes a Small Multidrug Resistance (SMR) family transporter, are located between two identical IS elements (IS3 family). Next to genes putatively involved in antibiotic resistance ICETn43716035 from C. litoralis KT71 also carries genes encoding a bacterial luciferase and a flavin reductase. ICETn43716140 from A. ebreus TPSY contains genes involved in metabolism (putative propanoate and butanoate metabolism) and a cluster encoding an ATP-Binding Cassette (ABC) transporter, an outer membrane protein and putative RNA modifying enzymes. The region rlxS-AccMod-TraMod of this element is highly similar (> 90% nucleotide identity) to the corresponding part of a partial element ΔICETn43716149 from A. denitrificans BC.
ICETn4371 Elements Coding for Unknown or Undefined Functions
R. solanacearum GMI1000 ICETn43716142 and MolK2 ICETn437161433 have AccMods with less defined genes. For the partial element ΔICETn43716150 from Acidovorax sp JS42 it is more difficult to describe the AccMod region as this element has no transfer module (to delineate the accessory genes). As mentioned above, numerous cases of ICETn4371-like elements have been observed in Alphaproteobacteria and the element ICETn43716139 in Xanthobacter autotrophicus Py2 has been shown as a representative. Although it does not contain an AccMod between rlxS and traG, it does contain numerous IS elements between int and TraMod. The spectrum of accessory genes in these Alphaprotobacterial elements and their location in the elements will need further study.
Transfer Genes and TraMod
Transfer genes that make up the transfer module are organized into two groups separated by the AccMod. The first group is composed of two genes, one coding for the TraF protease acting upon the pilus assembly protein TrbC,53 and the other coding for a RlxS (VirD2) relaxase.54,55 In fact, the traF gene likely belongs to a four-gene transcriptional unit parAparGgicCtraF with four base pairs overlaps. This organization, which links key functions involved in partition/replication and conjugational transfer, is conserved in almost all ICETn4371 including the Tn4371-like elements of the Alphaproteobacteria. It is not observed in the IncP/Ti plasmids and hence appears as a characteristic feature of ICETn4371. The ICETn4371 relaxases are similar to the RlxS (25%) of ICEMlSymR7A (Fig. 4) where this protein has been shown to be necessary for transfer and maintenance.34
The second part of the gene cluster involved in conjugational transfer is located downstream of the AccMod. This cluster begins with a gene coding for a putative transcriptional regulator protein TraR and a homolog of the coupling protein TraG. The latter, which is a putative DNA binding protein responsible for DNA transfer during conjugation,56 is similar to those in IncP plasmids. IncP plasmids are the best-studied broad host range plasmids, and thought to be among the most promiscuous of all plasmids known to-date. IncP plasmids are the subject of considerable research as they can spread across taxonomic barriers and therefore contribute to the rapid adaptation of various bacterial populations in natural and clinical environments.57 IncP plasmids, which replicate in the major branches of the Proteobacteria (Alpha, Beta and Gamma), are also known to have a transfer/mobilisation range that covers the Deltaproteobacteria58 and even extends far outside the Proteobacteria to the Actinobacteria,59 cyanobacteria and even yeasts.60 Also for Ti plasmids, which have a narrow replicative host range, the transfer range may be broader than expected although extensive studies remain to be done.61,62 IncP plasmids carry accessory genes involved in mercury resistance and degradation of xenobiotic organic compounds as well as antibiotic resistance genes and also share with many other broad host range plasmids the property to capture genes (also called "retrotransfer").63,64 Putatively, these gene dissemination features could be conserved, adapted and used by the ICETn4371 (maybe, for example, to capture AccMods). Interestingly, the gene order of two TraMod parts is also suggestive of an insertion of the AccMod into a primordial transfer module. The presence of transfer genes similar to plasmid genes exists in other ICE families. In the R391/SXT type of ICEs, the transfer genes (and a number of other R391/SXT core genes) are highly similar to those in IncA/C conjugative plasmids.13
The traG gene is followed by a group of genes coding for proteins (TrbBCDEJLFGI) with similarity to the mating-pair formation (mpf) apparatus of IncP and Ti plasmids (except for genes trbK and trbH, which are missing on the ICE) that are also related to the Type IV secretion system (Table 2).65 Together TraG and the TrbBCDEJLFGI proteins most likely mediate the transfer of ICETn4371 to recipient cells.66,67 In all the ICETn4371 elements the gene order is trbBCDEJLFGI, similar to the organization in ICEMlSymR7A (Fig. 4).43 Yet between traG and trbB lies a gene showing a strong identity with genes encoding repressors of the copG/metJ/arc/mnt family, which regulate the copy number of plasmids and phages and for which the trbR appellation is proposed. This copG-like gene very often overlaps by four base pairs with its neighbors traG and trbB including in Tn4371-like elements from Alphaproteobacteria. This may reflect a functional coupling between the partition/replication regulation and conjugational transfer as it may also be hypothesized with the parAG gicC traF operon in the StaMod.
Table 2.
Similarities/homologies between the transfer region of Tn4371 and other Type IV secretion systems.
A comparison of the Tra2 region of the IncP plasmid RP4, and the mating-pair formation apparatus of both R. pickettii 12J ICETn43716033 and ICEMlSymR7A can be seen in Figure 4. Protein similarities varied between 21-65% for ICETn43716033 and ICEMlSymR7A and ICETn43716033 and RP4 protein similarity varied between 11-38%. A similar study was performed with the VirB system from the Ti plasmid pNGR234a (CP000874) and ICETn43716033 however little homology was found.
Additional genes lacking in Ti and IncP plasmids can be found inserted between the traR and traG genes and the trbJ and trbL genes. These genes code for hypothetical lipoproteins, which are now annotated as gilB1 and gilB2 and that share some identity. These genes are found in most of the elements (Fig. 2) as well as the last gene gir, tentatively considered as involved in the excision process (see above).
Conclusion
Tn4371-related integrative and conjugative elements are found in a wide range of Alpha-, Beta- and Gammaproteobacteria from both clinical and environmental origin. These types of bacteria are known for their large metabolic repertoires and the ICETn4371 elements appear to be a source of adaptive functions. The recent attention to environmental microbiology, the incidence of chemical pollution and increase of nosocomial infections has contributed to reveal the role of this otherwise elusive family of mobile genetic elements in microbial evolution and adaptation to anthropogenic conditions.
Although, ICETn4371-related elements are present in Alpha-, Beta- and Gammaproteobacteria two major groups could be discerned. One group specific for the Beta- and Gammaproteobacteria, which characteristically carries its accessory genes between the relaxase (rlxS) and a transcriptional regulator (traR), while the second group is specific for the Alphaproteobacteria and the presence of accessory genes is scarce. These groups look quite exclusive as it would be expected from a taxonomic barrier. Still, the order of the transfer (trb) genes of both ICETn4371 groups is the same and similar to that of the Ti plasmid (virulence plasmid of Alphaproteobacterium Agrobacterium) (Table 2). In the Ti plasmid and ICETn4371 elements the trbJ, trbK and trbL genes lie further upstream of the trb operon, while these are the last genes of the IncP trb operon. This obvious translocation may suggest that the ancestral ICETn4371 was carried by a host most closely related to Alphaproteobacteria, which branched earlier in the tree of Proteobacteria than the Beta- and Gammaproteobacteria. Phylogenetic studies of the Type IV secretion/transfer systems may help to explore if these four types of elements, viz. ICETn4371 elements in Beta- and Gammaproteobacteria, ICEs in Alphaproteobacteria, Ti plasmids involved in plant-bacteria interactions, and broad host range IncP plasmids, indeed share a common evolutionary path, and to explore their relationship with integration vs. autonomy of mobile genetic elements.
The gene core of ICETn4371, which is composed of approximately 31 genes, harbours an int gene coding for a TBSSR, 13 genes in the StaMod and 17 genes involved in the TraMod. Among this group at least eight conserved genes are so far specific for ICETn4371 elements including four coding for putative lipoproteins (gilA1 and gilA2 in StaMod, gilB1 and gilB2 in TraMod), three coding for hypothetical proteins (gicA, gicB downstream parB, and gicC in parAGgicCtraF operon), and gir putatively involved in excision. Their association with the different modules and key functions in these modules suggest a unique role in the biology of ICETn4371 elements and justifies future genetic and functional studies.
The accessory gene module, which can be quite extensive ranging up to 70 kb, is always inserted into the TraMod (even if accessory genes are regularly found in the IntMod). Hereby it separates the relaxase (rlxS) from the transfer (trb) system and such a placement could be favorable for on the one hand maintaining the connection between the IntMod-StaMod and TraMod, and on the other be in control of the maximum accessory module insert size. In addition, the transfer function is transcriptionally coupled to the stabilization functions in a four-gene operon (parAGgicCtraF) and together with the distribution of the ICETn4371-specific conserved genes in both the StaMod and TraMod could act as additional regulatory aspects that allow the acquisition of an extensive baggage of accessory genes without the loss of its integration and transfer functions.
Acknowledgments
This work was supported by the European Space Agency (ESA-PRODEX) and the Belgian Science Policy (Belspo) through the COMICS project (C90356). Thanks to Pieter Monsieurs for constructing the heat map.
References
- 1.
- Springael D, Kreps S, Mergeay M. Identification of a catabolic transposon, Tn4371, carrying biphenyl and 4-chlorobiphenyl degradation genes in Alcaligenes eutrophus A5. J Bacteriol. 1993;175:1674–81. PMID:8383664. [PMC free article: PMC203961] [PubMed: 8383664]
- 2.
- Merlin C, Springael D, Toussaint A. Tn4371: A modular structure encoding a phage-like integrase, a Pseudomonas-like catabolic pathway, and RP4/Ti-like transfer functions. Plasmid. 1999;41:40–54. PMID:9887305 doi:10.1006/plas.1998.1375. [PubMed: 9887305]
- 3.
- Toussaint A, Merlin C, Monchy S, et al. The biphenyl- and 4-chlorobiphenyl-catabolic transposon Tn4371, a member of a new family of genomic islands related to IncP and Ti plasmids. Appl Environ Microbiol. 2003;69:4837–45. PMID:12902278 doi:10.1128/AEM.69.8.4837-4845.2003. [PMC free article: PMC169086] [PubMed: 12902278]
- 4.
- Ryan MP, Pembroke JT, Adley CC. Novel Tn4371-ICE like element in Ralstonia pickettii and genome mining for comparative elements. BMC Microbiol. 2009;9:242. PMID:19941653 doi:10.1186/1471-2180-9-242. [PMC free article: PMC2789088] [PubMed: 19941653]
- 5.
- Van Houdt R, Monchy S, Leys N, Mergeay M. New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria. Antonie van Leeuwenhoek. 2009;96:205–26. PMID:19390985 doi:10.1007/s10482-009-9345-4. [PubMed: 19390985]
- 6.
- Janssen PJ, Van Houdt R, Moors H, et al. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS ONE. 2010;5:e10433. PMID:20463976 doi:10.1371/journal.pone.0010433. [PMC free article: PMC2864759] [PubMed: 20463976]
- 7.
- Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155:376–86. PMID:15207870 doi:10.1016/j.resmic.2004.01.012. [PubMed: 15207870]
- 8.
- Franke AE, Clewell DB. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J Bacteriol. 1981;145:494–502. PMID:6257641. [PMC free article: PMC217299] [PubMed: 6257641]
- 9.
- Sen S, Oriel P. Transfer of transposon Tn916 from Bacillus subtilis to Thermus aquaticus. FEMS Microbiol Lett. 1990;55:131–4. PMID:2158473 doi:10.1111/j.1574-6968.1990.tb13849.x. [PubMed: 2158473]
- 10.
- Poyart C, Celli J, Trieu-Cuot P. Conjugative transposition of Tn916-related elements from Enterococcus faecalis to Escherichia coli and Pseudomonas fluorescens. Antimicrob Agents Chemother. 1995;39:500–6. PMID:7726521. [PMC free article: PMC162567] [PubMed: 7726521]
- 11.
- Roberts AP, Chandler M, Courvalin P, et al. Revised nomenclature for transposable genetic elements. Plasmid. 2008;60:167–73. PMID:18778731 doi:10.1016/j.plasmid.2008.08.001. [PMC free article: PMC3836210] [PubMed: 18778731]
- 12.
- Lechner M, Schmitt K, Bauer S, et al. Genomic island excisions in Bordetella petrii. BMC Microbiol. 2009;9:141. PMID:19615092 doi:10.1186/1471-2180-9-141. [PMC free article: PMC2717098] [PubMed: 19615092]
- 13.
- Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8:552–63. PMID:20601965 doi:10.1038/nrmicro2382. [PubMed: 20601965]
- 14.
- Marrero J, Waldor MK. The SXT/R391 family of integrative conjugative elements is composed of two exclusion groups. J Bacteriol. 2007;189:3302–5. PMID:17307849 doi:10.1128/JB.01902-06. [PMC free article: PMC1855829] [PubMed: 17307849]
- 15.
- Marrero J, Waldor MK. Determinants of entry exclusion within Eex and TraG are cytoplasmic. J Bacteriol. 2007;189:6469–73. PMID:17573467 doi:10.1128/JB.00522-07. [PMC free article: PMC1951900] [PubMed: 17573467]
- 16.
- Marrero J, Waldor MK. Interactions between inner membrane proteins in donor and recipient cells limit conjugal DNA transfer. Dev Cell. 2005;8:963–70. PMID:15935784 doi:10.1016/j.devcel.2005.05.004. [PubMed: 15935784]
- 17.
- Haase J, Kalkum M, Lanka E. TrbK, a small cytoplasmic membrane lipoprotein, functions in entry exclusion of the IncP alpha plasmid RP4. J Bacteriol. 1996;178:6720–9. PMID:8955288. [PMC free article: PMC178567] [PubMed: 8955288]
- 18.
- Creevey CJ, McInerney JO. Clann: investigating phylogenetic information through supertree analyses. Bioinformatics. 2005;21:390–2. PMID:15374874 doi:10.1093/bioinformatics/bti020. [PubMed: 15374874]
- 19.
- Jayaram M, Grainge I, Tribble G. In: Craig NL, Cragie R, Gellert M, Lambowitz AM, eds. Mobile DNA II. Washington, DC: ASM Press; 2002. Site-Specific recombination by the Flp protein of Saccharomyces cerevisiae; pp. 192–218.
- 20.
- Van Duyne GD. In: Craig NL, Cragie R, Gellert M, Lambowitz AM, eds. Mobile DNA II. Washington, DC: ASM Press; 2002. A structural view of tyrosine recombinase site-specific recombination; pp. 93–117.
- 21.
- Rice PA. In: Craig NL, Cragie R, Gellert M, Lambowitz AM, eds. Mobile DNA II. Washington, DC: ASM Press; 2002. Theme and variation in tyrosine recombinases: structure of a Flp-DNA complex; pp. 219–229.
- 22.
- Bouvier M, Demarre G, Mazel D. Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J. 2005;24:4356–67. PMID:16341091 doi:10.1038/sj.emboj.7600898. [PMC free article: PMC1356339] [PubMed: 16341091]
- 23.
- Dubois V, Debreyer C, Litvak S, Quentin C, Parissi V. A new in vitro strand transfer assay for monitoring bacterial class 1 integron recombinase IntI1 activity. PLoS ONE. 2007;2:e1315. PMID:18091989 doi:10.1371/journal.pone.0001315. [PMC free article: PMC2117344] [PubMed: 18091989]
- 24.
- Dubois V, Debreyer C, Quentin C, Parissi V. In vitro recombination catalyzed by bacterial class 1 integron integrase IntI1 involves cooperative binding and specific oligomeric intermediates. PLoS ONE. 2009;4:e5228. PMID:19381299 doi:10.1371/journal.pone.0005228. [PMC free article: PMC2668188] [PubMed: 19381299]
- 25.
- Cambray G, Guerout AM, Mazel D. Integrons. Annu Rev Genet. 2010;44:141–66. PMID:20707672 doi:10.1146/annurev-genet-102209-163504. [PubMed: 20707672]
- 26.
- Boyd EF, Almagro-Moreno S, Parent MA. Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 2009;17:47–53. PMID:19162481 doi:10.1016/j.tim.2008.11.003. [PubMed: 19162481]
- 27.
- Hallet B, Van hoof V, Cornet F. In: Funnell B, Phillips G, eds. Plasmid Biology. Washington, DC: ASM Press; 2004. DNA site-specific resolution systems; pp. 145–179.
- 28.
- Klimke W, Agarwala R, Badretdin A, et al. The National Center for Biotechnology Information's Protein Clusters Database. Nucleic Acids Res. 2009;37:D216-23. PMID:18940865 doi:10.1093/nar/gkn734. [PMC free article: PMC2686591] [PubMed: 18940865]
- 29.
- Wang H, Mullany P. The large resolvase TndX is required and sufficient for integration and excision of derivatives of the novel conjugative transposon Tn5397. J Bacteriol. 2000;182:6577–83. PMID:11073898 doi:10.1128/JB.182.23.6577-6583.2000. [PMC free article: PMC111396] [PubMed: 11073898]
- 30.
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–7. PMID:14960472 doi:10.1093/bioinformatics/btg430. [PubMed: 14960472]
- 31.
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. PMID:15034147 doi:10.1093/nar/gkh340. [PMC free article: PMC390337] [PubMed: 15034147]
- 32.
- Lewis JA, Hatfull GF. Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 2001;29:2205–16. PMID:11376138 doi:10.1093/nar/29.11.2205. [PMC free article: PMC55702] [PubMed: 11376138]
- 33.
- ElAntak L, Ansaldi M, Guerlesquin F, Mejean V, Morelli X. Structural and genetic analyses reveal a key role in prophage excision for the TorI response regulator inhibitor. J Biol Chem. 2005;280:36802–8. PMID:16079126 doi:10.1074/jbc.M507409200. [PubMed: 16079126]
- 34.
- Ramsay JP, Sullivan JT, Stuart GS, Lamont IL, Ronson CW. Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol Microbiol. 2006;62:723–34. PMID:17076666 doi:10.1111/j.1365-2958.2006.05396.x. [PubMed: 17076666]
- 35.
- Ramsay JP, Sullivan JT, Jambari N, et al. A LuxRI-family regulatory system controls excision and transfer of the Mesorhizobium loti strain R7A symbiosis island by activating expression of two conserved hypothetical genes. Mol Microbiol. 2009;73:1141–55. PMID:19682258 doi:10.1111/j.1365-2958.2009.06843.x. [PubMed: 19682258]
- 36.
- Austin S, Ziese M, Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell. 1981;25:729–36. PMID:7026049 doi:10.1016/0092-8674(81)90180-X. [PubMed: 7026049]
- 37.
- Gerlitz M, Hrabak O, Schwab H. Partitioning of broad-host-range plasmid RP4 is a complex system involving site-specific recombination. J Bacteriol. 1990;172:6194–203. PMID:2172207. [PMC free article: PMC526800] [PubMed: 2172207]
- 38.
- Bignell C, Thomas CM. The bacterial ParA-ParB partitioning proteins. J Biotechnol. 2001;91:1–34. PMID:11522360 doi:10.1016/S0168-1656(01)00293-0. [PubMed: 11522360]
- 39.
- Gerdes K, Moller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol. 2000;37:455–66. PMID:10931339 doi:10.1046/j.1365-2958.2000.01975.x. [PubMed: 10931339]
- 40.
- Burrus V, Pavlovic G, Decaris B, Guedon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46:601–10. PMID:12410819 doi:10.1046/j.1365-2958.2002.03191.x. [PubMed: 12410819]
- 41.
- Wozniak RA, Waldor MK. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009;5:e1000439. PMID:19325886 doi:10.1371/journal.pgen.1000439.
- 42.
- Qiu X, Gurkar AU, Lory S. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2006;103:19830–5. PMID:17179047 doi:10.1073/pnas.0606810104. [PMC free article: PMC1750864] [PubMed: 17179047]
- 43.
- Sullivan JT, Trzebiatowski JR, Cruickshank RW, et al. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J Bacteriol. 2002;184:3086–95. PMID:12003951 doi:10.1128/JB.184.11.3086-3095.2002. [PMC free article: PMC135072] [PubMed: 12003951]
- 44.
- Marenda M, Barbe V, Gourgues G, Mangenot S, Sagne E, Citti C. A new integrative conjugative element occurs in Mycoplasma agalactiae as chromosomal and free circular forms. J Bacteriol. 2006;188:4137–41. PMID:16707706 doi:10.1128/JB.00114-06. [PMC free article: PMC1482908] [PubMed: 16707706]
- 45.
- Vallenet D, Labarre L, Rouy Z, et al. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 2006;34:53–65. PMID:16407324 doi:10.1093/nar/gkj406. [PMC free article: PMC1326237] [PubMed: 16407324]
- 46.
- Tseng TT, Gratwick KS, Kollman J, et al. The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–25. PMID:10941792. [PubMed: 10941792]
- 47.
- Furukawa H, Tsay JT, Jackowski S, Takamura Y, Rock CO. Thiolactomycin resistance in Escherichia coli is associated with the multidrug resistance efflux pump encoded by emrAB. J Bacteriol. 1993;175:3723–9. PMID:8509326. [PMC free article: PMC204787] [PubMed: 8509326]
- 48.
- Altendorf K, Gassel M, Puppe W, et al. Structure and function of the Kdp-ATPase of Escherichia coli. Acta Physiol Scand Suppl. 1998;643:137–46. PMID:9789555. [PubMed: 9789555]
- 49.
- Altendorf K, Siebers A, Epstein W. The KDP ATPase of Escherichia coli. Ann N Y Acad Sci. 1992;671:228–43. PMID:1288322 doi:10.1111/j.1749-6632.1992.tb43799.x. [PubMed: 1288322]
- 50.
- Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev. 2003;27:313–39. PMID:12829273 doi:10.1016/S0168-6445(03)00048-2. [PubMed: 12829273]
- 51.
- Hundertmark M, Hincha DK. LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics. 2008;9:118. PMID:18318901 doi:10.1186/1471-2164-9-118. [PMC free article: PMC2292704] [PubMed: 18318901]
- 52.
- Mijnendonckx K, Provoost A, Monsieurs P, et al. Insertion sequence elements in Cupriavidus metallidurans CH34: Distribution and role in adaptation. Plasmid. 2011;65:193–203. PMID:21185859 DOI:doi:10.1016/j.plasmid.2010.12.006. [PubMed: 21185859]
- 53.
- Haase J, Lanka E. A specific protease encoded by the conjugative DNA transfer systems of IncP and Ti plasmids is essential for pilus synthesis. J Bacteriol. 1997;179:5728–35. PMID:9294428. [PMC free article: PMC179460] [PubMed: 9294428]
- 54.
- Byrd DR, Matson SW. Nicking by transesterification: the reaction catalysed by a relaxase. Mol Microbiol. 1997;25:1011–22. PMID:9350859 doi:10.1046/j.1365-2958.1997.5241885.x. [PubMed: 9350859]
- 55.
- Lanka E, Wilkins BM. DNA processing reactions in bacterial conjugation. Annu Rev Biochem. 1995;64:141–69. PMID:7574478 doi:10.1146/annurev.bi.64.070195.001041. [PubMed: 7574478]
- 56.
- Gomis-Rüth FX, de la Cruz F, Coll M. Structure and role of coupling proteins in conjugal DNA transfer. Res Microbiol. 2002;153:199–204. PMID:12066890 doi:10.1016/S0923-2508(02)01313-X. [PubMed: 12066890]
- 57.
- Thomas CM. Amsterdam: Harwood Academic Publishers; 2000. The Horizontal Gene Pool—Bacterial Plasmids and Gene Spread.
- 58.
- Powell B, Mergeay M, Christofi N. Transfer of broad host-range plasmids to sulphate-reducing bacteria. FEMS Microbiol Lett. 1989;59:269–73. doi:10.1111/j.1574-6968.1989.tb03123.x.
- 59.
- Gormley EP, Davies J. Transfer of plasmid RSF1010 by conjugation from Escherichia coli to Streptomyces lividans and Mycobacterium smegmatis. J Bacteriol. 1991;173:6705–8. PMID:1657866. [PMC free article: PMC209018] [PubMed: 1657866]
- 60.
- Heinemann JA, Sprague GF Jr. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989;340:205–9. PMID:2666856 doi:10.1038/340205a0. [PubMed: 2666856]
- 61.
- Sprinzl M, Geider K. Transfer of the Ti plasmid from Agrobacterium tumefaciens into Escherichia coli cells. J Gen Microbiol. 1988;134:413–24. PMID:3049936. [PubMed: 3049936]
- 62.
- Farrand SK. In: Clewell DB, ed. Bacterial Conjugation. New York: Plenum Press; 1993. Conjugal transfer of Agrobacterium plasmids; pp. 255–291.
- 63.
- Szpirer C, Top E, Couturier M, Mergeay M. Retrotransfer or gene capture: a feature of conjugative plasmids, with ecological and evolutionary significance. Microbiology. 1999;145:3321–9. PMID:10627031. [PubMed: 10627031]
- 64.
- Haines AS, Akhtar P, Stephens ER, et al. Plasmids from freshwater environments capable of IncQ retrotransfer are diverse and include pQKH54, a new IncP-1 subgroup archetype. Microbiology. 2006;152:2689–701. PMID:16946264 doi:10.1099/mic.0.28941-0. [PubMed: 16946264]
- 65.
- Li PL, Everhart DM, Farrand SK. Genetic and sequence analysis of the pTiC58 trb locus, encoding a mating-pair formation system related to members of the type IV secretion family. J Bacteriol. 1998;180:6164–72. PMID:9829924. [PMC free article: PMC107700] [PubMed: 9829924]
- 66.
- Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. PMID:12855161 doi:10.1016/S0378-1097(03)00430-0. [PubMed: 12855161]
- 67.
- Schröder G, Lanka E. The mating pair formation system of conjugative plasmids-A versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54:1–25. PMID:15907535 doi:10.1016/j.plasmid.2005.02.001. [PubMed: 15907535]
- The Tn4371 ICE Family of Bacterial Mobile Genetic Elements - Madame Curie Biosci...The Tn4371 ICE Family of Bacterial Mobile Genetic Elements - Madame Curie Bioscience Database
- Idiopathic Parkinson's Disease: Staging an α-Synucleinopathy with a Predictable ...Idiopathic Parkinson's Disease: Staging an α-Synucleinopathy with a Predictable Pathoanatomy - Madame Curie Bioscience Database
- The Role of Heat Shock Proteins during Neurodegeneration in Alzheimer's, Parkins...The Role of Heat Shock Proteins during Neurodegeneration in Alzheimer's, Parkinson's and Huntington's Disease - Madame Curie Bioscience Database
- Abl Family Kinases in Mammalian Development - Madame Curie Bioscience DatabaseAbl Family Kinases in Mammalian Development - Madame Curie Bioscience Database
- Cell-Cell Channels and Their Implications for Cell Theory - Madame Curie Bioscie...Cell-Cell Channels and Their Implications for Cell Theory - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...