NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
RNA binding proteins (RBPs) are involved in each step of RNA metabolism. Most of them are composed of small RNA binding domains (RBDs) that are needed for their recruitment to specific RNA targets. The mode of RNA recognition of these RBDs has been studied by structural biologists for more than 20 years and seems to be highly versatile. In this chapter we review the current structural knowledge about RNA recognition by the four main RBD families, namely RNA recognition motifs (RRMs), zinc fingers, KH domains and double-stranded RNA binding motifs (dsRBMs) detailing how structural data have brought information essential for a better understanding of RBP functions.
INTRODUCTION
This book focuses on the extreme variety of functions carried by RNA binding proteins (RBPs). The association of RBPs with RNA transcripts begins during transcription. Some of these early-binding RBPs remain bound to the RNA until they are degraded, whereas others recognize and transiently bind to RNA at later stages for specific processes such as splicing, processing, transport and localization.1 Some RBPs also function as RNA chaperones2 by helping the RNA, which is initially single-stranded, to form various secondary or tertiary structures. When folded, these structured RNAs, together with specific RNA sequences, act as a signal for other RBPs that mediate gene regulation. Most of RNA binding proteins contain several domains including different types of RNA binding motifs, very often in multiple copies (Fig. 1), which recognize RNA sequence specifically.
Here, we review our current structural understanding of protein—RNA recognition mediated by four RNA-binding domains, the RNA recognition motif (RRM), the zinc-finger domain, the KH domain and the double-stranded RNA-binding motif (dsRBM). We discuss how these four small domains recognize RNA. Some bind single-stranded RNA by direct readout of the primary sequence, whereas others recognize primarily the shape of the RNA or both the sequence and the shape. This chapter shows how, within the past 15 years, structural biology revealed the highly versatile mode of protein-RNA interactions and contributed to explain their importance for RBP functions.
RNA RECOGNITION MOTIFS (RRMs)
The RNA-recognition motif (RRM), also known as RBD (RNA binding domain) or RNP (ribonucleoprotein domain) is the most abundant RNA-binding domain in higher vertebrates (this motif is present in about 0.5%-1% of human genes)3 and is the most extensively studied RNA-binding domain, both in terms of structure and biochemistry.4 Although this domain was also shown interacting with DNA and protein partners, we focus here on its role as an RNA binding domain. Typically, an RRM can be recognized at the primary sequence level as a 90 amino acids long domain containing two conserved sequences of eight and six amino-acids, called RNP1 and RNP2, respectively (Fig. 2A). RRM adopts a typical β1α1β2β3α2β4 topology that forms a four-stranded β-sheet packed against two α-helices (Fig. 2B). The RNP1 and RNP2 sequences located in the two central β-strands β3 and β1 of the domain, respectively, expose three conserved aromatic residues on the surface of the β-sheet which form the primary RNA binding surface. Lack of the presence of most if not all of these aromatic residues led to the definition of several subclass of RRMs like the quasi-RRM (qRRM), the pseudo-RRM (ΨRRM) or U2AF Homology Motifs (UHM).5
Since the first structure of an RRM-RNA complex in 1994 by Nagai and coworkers,6 more than 30 high-resolution structures of RRM-RNA complexes have been determined either by X-ray crystallography6-19 or NMR spectroscopy20-33 (Table 1). This fairly large ensemble of available structures revealed common features between the complexes and also many surprises.
Table 1.
Structures of RRMs solved in complex with RNA. The protein domains and the method used for the structure determination are indicated with the corresponding PDB number (http://www.rcsb.org/pdb/home/home.do).
A Simple Fold Binding a Large Panel of RNA Sequences and Structures
Binding the β-Sheet Surface
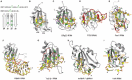
As maybe expected from sequence conservation, the four-stranded β-sheet constitutes the primary and most common RNA binding surface of an RRM. Typically, the three conserved aromatic side-chains located in the conserved sequences RNP1 (in β3-strand) and RNP2 (in β1-strand) (Fig. 2A), accommodate two nucleotides as follows: the bases of the 5′ and of the 3′ nucleotides stack on an aromatic ring located in β1 (position 2 of RNP2) and in β3 (position 5 of RNP1), respectively (Fig. 2B). The third aromatic ring located in β3 (position 3 of RNP1) is found often inserted between the two sugar rings of the dinucleotide (Fig. 2B). The RRM β-sheet surface is therefore a perfect platform to interact with two consecutive single-stranded RNA nucleotides. Three to four nucleotides are usually accommodated on the β-sheet, via the presence of aromatic rings or other planar side-chains (Arg, Asn, Asp, His) on the other β-strands as shown in the RRM structure of hnRNPA1 (Fig. 2B) or SRp20 (Fig. 2C). If these aromatic rings and planar side-chains provide RNA binding affinity, they do not achieve sequence-specificity. RNA sequence-specificity by RRMs is achieved by side-chains present on the β-sheet surface but also by the main-chain of the residues immediately C-terminal to the RRM (i.e., the few residues following strand β4) as shown in the case of the RRM of hnRNPA1 (Fig. 2B) or SRp20 (Fig. 2C). This mode of binding allows a binding affinity in the micromolar range and a certain bias in sequence-specificity since the involvement of the main-chain in binding results in a binding preference of the canonical RRM for a W-G dinucleotide (W can be A or C).34 Although most of RRMs use this basic mode of binding, RRM evolved to be capable of binding a large repertoire of sequences by using parts of the domain outside the strict β-sheet surface.
The Extension of the β-Sheet Surface
One way to accommodate more nucleotides and therefore achieve higher affinity is to extend the β-sheet surface of the RRM. To date, two RRMs were shown to have such extended surface, the RRM2 and RRM3 of PTB which both contain a fifth β-strand anti parallel to β2 (Fig. 2D). The presence of this additional strand allowed the binding of one (RRM2) or two (RRM3) additional nucleotides.28 A second approach selected across evolution was to juxtapose two consecutive RRMs as in PAPB where β2 of RRM2 interacts with β4 of RRM1 creating a continuous surface that can accommodate eight adenines7 (Fig. 3).
The Critical Role of Protein Loops to Extend RNA Binding
In other RRMs, the loops connecting the β-strands or the β-strands to the α-helices of the RRMs can be involved in RNA recognition. The most spectacular example in this context is probably the RRM of Fox-1 where the β1-α1 and α2-β4 loops of human Fox-1 RRM22 are involved in the binding of the first four nucleotides of the sequence 5′-UGCAUGU-3′. In particular, one Phenylalanine located in the β1-α1 loop stacks with the first two RNA nucleotides, whereas the last three nucleotides are recognized in a canonical manner by the β-sheet of the RRM (Fig. 2E). This structure nicely shows how nonconserved parts of the RRM can be used to extend to more than double of the canonical binding surface. As a result Fox-1 RRM binds the heptamer 5′-UGCAUGU-3′ with subnanomolar affinity.
In addition to be involved in the specific binding of RNA sequences, RRM loops can also be responsible for the recognition of the RNA shape. This is probably best illustrated by the structure of RBMY RRM in complex with an RNA stem-loop, where all residues of the β2-β3 loop insert into the RNA major groove30 of the RNA helix while the β-sheet specifically recognizes the RNA loop (Fig. 2F). Similarly, the β2-β3 loop of the N-terminal RRM of U1A or of U2B′′ is also crucial to recognize a stem-loop6,14 or an internal loop.35
The Use of RRM N- and C-Terminal Regions
The N- and C-terminal regions, outside the RRM, are usually poorly ordered in the isolated domains with a few exceptions where they can adopt a secondary structure. For example, in the structures of the La C-terminal RRM,36 the N-terminal RRM of U1A37 of CstF-64 RRM38 or more recently in the qRRM of hnRNP F,24 the C-terminus forms a α-helix that covers the β-sheet surface of the RRM preventing its access to RNA. However, in an increasing number of RRMs, the regions outside the predicted RRMs have proved to be of crucial importance to significantly enhance RNA binding affinity and to play a role in sequence-specificity. Several structures show that the N-terminal (CUG-BP1, CBP20),31,39 the C-terminal (PABP and PTB)7,28 or both the N- and C-termini (Tra2-β1)23,32 regions of the RRM can be used to directly interact with the targeted RNA. In this latter example, both N- and C-terminal regions of Tra2-β1 RRM are found unstructured in the free RRM and become ordered only upon RNA binding. Both terminal regions interact with RNA and cross each other23,32 (Fig. 2G). Amino acids from these extremities and from the β-sheet participate to the specific recognition of the 5′-AGAA-3′ sequence (Fig. 2G). This positioning of two termini induced upon RNA binding in Tra2-β1 could be functionally important as it could explain how Tra2-β1 recruits two additional proteins, hnRNPG and SRp30c, on SMN (survival of motoneuron) exon 723 in order to increase its splicing.40,41
The qRRM, an RRM Binding RNA without Using the β-Sheet Surface
In contrast to the RRMs discussed above which always use the canonical β-sheet surface, the qRRMs of hnRNP F bind RNA in a different way. Indeed, the β1/α1, β2/β3 and α2/β4 loops, but not the β-sheet, interact with RNA.24 Due to the lack of conserved aromatic residues in RNP1 and RNP2, the three RRMs of hnRNP F have been renamed qRRMs for quasi RRMs.42 The recent structures of the three RRMs of hnRNP F in complex with a G-tract of three guanines show that each qRRM binds RNA in an identical manner. The three guanines adopt a compact conformation surrounded by three conserved residues belonging to protein loops that are stacking with each guanine base (two aromatics and one arginine). The qRRMs appear to encage the G-tract24 (Fig. 2H). This mode of binding appears conserved among qRRMs, since the side-chains involved in sequence-specific recognition of the G-tract are conserved among identified qRRMs.24 By sequestering in this manner G triplets, it was proposed that hnRNP F could regulate splicing by maintaining G-rich sequences in a single stranded conformation, therefore preventing RNA to fold into a secondary structure.24
The RRM, an Extraordinary Plastic RNA Binding Module
Altogether, these structural investigations showed that the interaction of RRM with RNA is not restricted to the β-sheet surface and that the loops (especially loops 1, 3 and 5) and the two termini appear to be equally important to the β-sheet for binding RNA with high affinity and sequence-specificity. Although, the canonical β-sheet surface appears to interact preferably with a C/A-G sequence, RRMs are found binding almost any types of sequences and secondary structures showing the extraordinary plasticity of this RNA binding domain. In this context, the recent structure of an RRM of the bacterial DEAD-box helicase YxiN bound to RNA perfectly illustrates the extraordinary plasticity of an RRM, since this RRM in order to recognize a three-way RNA junction requires the involvement of almost all parts of the domain: β-sheet, loops and even the helices (Fig. 2I).9
When Multiple RRMs Bind RNA
Proteins containing multiple RRMs seem to be the rule more than the exception. Often the RRMs within one protein show high similarity in primary sequence and therefore the RNA binding specificity of each RRM can be almost identical (the three RRMs of hnRNP F or the two RRMs of PABP or hnRNP A1) or similar (the four RRMs of PTB or the three RRMs of CUG-BP). Two RRMs within one protein can be also highly dissimilar in sequence and function as shown for the RRMs of U1A, U2B′′, U2AF65 and U2AF35 or of several SR proteins.4 Yet, the ways multiple RRMs are used by each proteins to bind RNA seem to differ drastically.
Multiple RRMs to Achieve Higher RNA Binding Affinity and Sequence-Specificity
As one would expect, similarly to what was shown for DNA binding modules, having RRMs in tandem allows higher RNA binding affinity and sequence-specificity compared to single RRM if both domains contribute to RNA binding. This has been nicely illustrated with the structures of several tandem RRMs bound to RNA, namely Sex-lethal,8 PABP,7 HuD,18 nucleolin,20 or more recently Hrp1.29 In all five complexes, the two RRMs bind synergistically a continuous single-stranded RNA sequence or stem-loop. The β-sheet of both RRMs is the primary binding surface, however the short (around ten amino-acids) interdomain linker between the two RRMs plays a key role for the RNA recognition. In all five proteins, the interdomain linker is flexible in the free state to become well ordered upon RNA binding. This further illustrates the importance of the N- and C-terminal extension of RRMs for RNA recognition. In all complexes, RRM2 and RRM1 bind the upstream and downstream RNA sequence, respectively and for the complexes of Hrp1,29 HuD18 and Sex-lethal8 the orientation of the two RRMs is almost identical, the two RRM forming a cleft (Fig. 3A). In PABP,7 the β-sheets of both domains are adjacent forming more a large RNA binding platform (Fig. 3B), while in nucleolin,20 two RRMs sandwiched the RNA (Fig. 3C).
Multiple RRMs to Affect the RNA Topology
In PTB or in hnRNPA1, tandem RRMs interact already in the free state positioning these consecutive domains in such a way that they cannot bind adjacent sequences. The structures of the complexes with single-stranded RNA and DNA for PTB RRM34 and hnRNPA1 RRM12, respectively, indeed showed that such orientations of the RRMs could induce the formation of RNA loops between the two binding sites of each RRM (Fig. 3D). This mode of binding could explain the mode of action of both proteins as repressor of splicing.28,43
When Multiple RRMs Bind Independently, What is the Function?
In many proteins containing multiple RRMs, the RRMs do not interact with each other and appear to bind RNA independently from each other. In PTB for example, RRM1 and RRM2 are clearly independent from the other RRMs and are separated by long flexible linkers. The same is true for hnRNP F qRRMs (Dominguez and Allain, unpublished). Within each hnRNP protein, each RRM binds very similar sequences (UCU for PTB RRMs and GGG for hnRNP F qRRMs) with micromolar affinity. Part of the function for this independent binding of each RRM might be to increase the chance for the protein to encounter their specific binding sequence. Having long flexible linkers between the RRMs allow the protein to span large volumes and therefore increase the probability to find an accessible RNA binding sequence. The same might hold true for two other splicing factors U2AF65 and CUG-BP for which each RRM appears to bind independently U-rich and GU-rich sequences, respectively.15,16,31
When Multi-RRM Proteins Compete Functionally
In light of the three mode of RNA binding described above, it is interesting to compare Sex-lethal, PTB and U2AF both structurally and functionally since these three multi-RRMs bind all the 3′ splice-site pyrimidine-tract yet with a different mode of binding. The fact that all three proteins can function at the same site when the 3′ splice-site sequence is optimal for the binding for each protein44 although their RNA binding mode is very different argues that a precise binding place dictates the function more than the binding mode. If Sex-lethal has the advantage of binding with higher affinity due to the cooperativity of binding between the two RRMs, U2AF and PTB although they bind more weakly, have the advantage over Sex-lethal to be less stringent with the RNA target they can bind both in term of sequence and length. Similar competitions between RNA binding proteins using a different mode of binding is likely to be frequent in RNA biology which render the molecular mechanisms of posttranscriptional gene regulation difficult to decipher or to model.
THE ZINC FINGER DOMAIN
Zinc finger (ZnF) is another domain found in RNA binding proteins. In a single RBP this motif can be found alone, as a repeated domain or even in combination with other types of RBDs (Fig. 1). A classical zinc finger is about 30 amino acids long and displays a ββα protein fold in which a β-hairpin and an α-helix are pinned together by a Zn2+ ion. These domains are classified depending on the amino acids that interact with this ion (e.g., CCHH, CCCH or CCCC) and were initially described as DNA binding motifs, the CCHH-type being the most frequent one. They were found interacting specifically with dsDNA bases located in major grooves via side chains of residues present in their α-helix.45 More recently, zinc fingers have also been shown binding RNA molecules. It therefore raised a series of questions. How such a small domain accommodates RNA? Is it able to recognize specifically a RNA sequence? How versatile is this interaction? In this section, referring to the few structures of zinc fingers solved in complex with RNA46-54 (Table 2), we give a short overview of what is known about the RNA binding mode of these surprising RNA binding motifs.
Table 2.
Structures of zinc fingers solved in complex with RNA. The protein domains and the method used for the structure determination are indicated with the corresponding PDB number (http://www.rcsb.org/pdb/home/home.do) .
The High Diversity of Interaction of Zinc Fingers with RNA
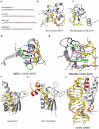
The structure of TFIIIA zinc fingers was among the first to be solved in complex with RNA. TFIIIA is a transcription factor that was shown to be involved in the transcription of eukaryotic ribosomal 5S RNA.55 This protein contains nine CCHH zinc fingers and can either bind DNA or RNA molecules.56 The crystal structure of TFIIIA ZnF4 to 6 in complex with a minimal folded version of the 5S rRNA53 revealed that zinc fingers were able to interact with the backbone of a RNA double-helix and could even specifically recognize individual accessible bases. However, at that time, it was not clear whether these domains could drive proteins to specific ssRNA sequences, as RRMs do. Such evidence came later from other types of zinc fingers (CCCH, CCHC and CCCC) for which the structure was primarily solved in complex with single stranded RNAs (Fig. 4A–E).
Tis11d, a member of the tristetrapolin (TTP) protein family, contains two CCCH zinc fingers (Fig. 1). This protein is involved in the control of the inflammatory response and induces the degradation of mRNAs that contain an AU-rich element (ARE) in their 3′-UTR.57 In order to understand how this recognition occurs, the structure of the two Tis11d zinc fingers was solved by NMR in complex with the RNA sequence 5′-UUAUUUAUU-3′.50 This structural study shows that an UAU motif is specifically recognized by each domain through hydrogen bonds mediated primarily by protein backbone atoms to Watson-Crick edges of the bases (Fig. 4B). A similar mode of interaction is used by CCHC zinc fingers of HIV-1 nucleocapsid protein (NC) to recognize a different motif.47 As illustrated in the Figure 4C, the ZnF1 of NC interacts specifically with a 5′-AG-3′ dinucleotide. These structures demonstrated the ability of zinc fingers to recognize different RNA sequences. However, this RNA recognition mode using almost exclusively the backbone of the protein is not the only way for zinc fingers to specifically bind RNA. The next two examples reveal that these domains are in fact able to bind different RNA sequences with an unexpected diversity of interactions.
MBNL1 (Muscleblind-like 1) contains four CCCH zinc fingers (Fig. 1) and was also shown to bind RNA. This tissue-specific alternative splicing regulator was proposed to promote muscle differentiation.58 Understanding the mode of action of this protein is a current challenge since its inactivation is in part responsible for leading to the myotonic dystrophy disease. A crystal structure was solved showing that 5′-GC-3′ and 5′-GCU-3′ RNA motifs are specifically recognized by MBNL1 ZnF3 and 4, respectively, with a nanomolar affinity. As for most of the proteins that contain several RNA binding domains, the mode of RNA recognition is very similar for these two zinc fingers. As illustrated in Figure 4D with the ZnF3, this sequence-specific recognition is mediated by stacking interactions and several hydrogen bonds involving main chains and, contrary to Tis11d, also side chains of the protein. A particularity of the MBNL1-RNA complex is that two cysteines, (Cys185 and Cys200) co-ordinating zinc atom, also interact with RNA (Fig. 4D).
The structures of Tis11d and MBNL1 illustrate how two CCCH ZnF containing proteins can specifically recognize different RNA sequences, namely UAU or GC(U) using two distinct modes of interaction. However, the opposite is also observed since several proteins sharing sequence homologies in their zinc finger domains were found interacting with the same RNA sequence. An example is presented below with the atypical ZRANB2 zinc finger family.
ZRANB2 is a human splicing factor that contains two RanBP2-type ("CCCC") zinc finger domains followed by a RS domain (Fig. 1). As described in the chapter by Hertel et al in this volume the RS domains are characterized by a repetition of Arg-Ser dipeptides and are mainly involved in protein-protein interactions. The RanBP2-type domain binds RNA and is defined by the consensus sequence W-X-C-X2-4-C-X3-N-X6-C-X2-C. This domain adopts an unusual fold that comprises two short β-hairpins sandwiching a central tryptophan residue and a single zinc ion co-ordinated by four conserved cysteines59 (Fig. 4E). A crystal structure of the two ZRANB2 ZnFs in complex with the RNA sequence 5′-AGGUAA-3′ was determined recently.52 A structural particularity of this RNA-protein complex is the guanine-Trp79-guanine "ladder" formation adopted by a continuous stacking of these three residues (Fig. 4E). The three consecutive bases G2, G3 and U4 are specifically recognized by formation of hydrogen bonds involving protein side chains (Asn76, Arg81, Arg82 and Asn86), backbone groups (Val77 carbonyl and Trp79 amide) and water-mediated hydrogen bonds (Asp68 and Ala80) (Fig. 4E). These amino acids are mainly located in ZRANB2 loops, especially the one located at the C-terminal extremity of the first β-hairpin (Fig. 4E). Based on these structural data and biochemical studies, six other ZRANB2 ZnF containing proteins involved in mRNA processing could be identified in humans as binding a GGU motif in ssRNAs with micromolar affinities.60
Which Functions for Zinc Fingers?
Structural data did not only help to better understand the specificity of interaction of these RNA binding domains and identify new putative binding motifs for ZnF binding proteins, they also helped to find new possible functions for these factors. For ZRANB2, based on functional data and the strong homology between its binding sequence evidenced by the structure and the 5′ splice-site sequences, the authors suggested that this protein might interact with a subset of 5′ splice-sites preventing their recognition by the spliceosome.52 Also based on structural data, the anti-parallel orientation observed for the RNA molecules bound by the two zinc fingers of MBNL1 and the location of its binding sites on natural targets suggested that this protein could induce a looping of the RNA, as observed for PTB RRM34, blocking the 3′ splice-site recognition by U2 snRNP and resulting in exon skipping.54
In this section, we showed that as described for RRMs, zinc fingers are able to specifically bind single-stranded RNA with affinities ranging from nanomolar to the micromolar, using primarily hydrogen bond and aromatic-base stacking interactions. However, the amino acids involved in RNA interaction are not primarily located in the β-strands like for most of RRMs but rather embedded in the protein loops and α-helices. Another characteristic of these domains is their ability to adopt different folds in order to be able to recognize different sequences using their main chains. Due to the apparent wide diversity of RNA-ZnF interactions, more structures are now needed to better understand and classify the different modes of RNA recognition of these small RNA binding domains.
THE KH DOMAIN
The hnRNP K homology (KH) domain is approximately 70 amino acids long. It is found in proteins with different functions including splicing, transcriptional regulation and translational control. Two versions of the KH fold have been reported, the types I and II found in eukaryotic and prokaryotic proteins, respectively. The type I has a βααββα topology and is characterized by a β-sheet composed of three antiparallel β-strands packed against three α-helices.61,62 The β1- and β2-strands are parallel to each other and the β3-strand is antiparallel to both. In addition, a "GXXG loop" containing the (I/L/V)-I-G-X-X-G-X-X-(I/L/V) conserved motif, located between the α1 and α2 helices, and a β2-β3 loop variable in length (3 to over 60 amino acids) and sequence, are also found in this motif (Fig. 4F). The KH type II fold differs from the type I by a αββααβ topology and a characteristic β-sheet in which the central strand (β2) is parallel to β3 and antiparallel to β1.61,62 Although both KH motif folds are known for interacting with RNA or ssDNA targets, only few structures of these domains bound to nucleic acid molecules have been deposed in the Protein Data Bank63-69 (Table 3) and most of them concern the eukaryotic type I KH domain. Therefore, we will essentially focus on this type of KH fold in this section.
Table 3.
Structures of KH domains solved in complex with RNA. The protein domains and the method used for the structure determination are indicated with the corresponding PDB number (http://www.rcsb.org/pdb/home/home.do) .
KH Domains Bind Four Nucleotides
KH domains have been shown interacting with their nucleic acid targets using common features. Typically, the single-stranded RNA or DNA molecule is mostly bound by an extended RNA binding surface including the α1 and α2 helices linked by the GXXG loop on one side and the β-sheet and the variable loop on the other side.62 Together, they form a binding cleft that usually accommodates four bases (Fig. 4F). As an example, we describe the mode of interaction of the KH3 domain of Nova2 (Neuro-oncological ventral antigen 2) with RNA. This tissue-specific alternative splicing factor is highly expressed in the neocortex and hippocampus where it regulates the alternative splicing of transcripts coding for proteins having specific functions in brain.70 Nova2 contains three KH domains of type I (Fig. 1). The crystal structure of the KH3 domain in complex with an in vitro selected stem-loop RNA shows that this protein interacts with the single stranded 5′-UCAC-3′ sequence located in the loop67 (Fig. 4F). U12 is specifically and indirectly recognized by two water molecules forming hydrogen bonds with the Lys23 and the Arg75 located in the GKGG protein loop and the α3 helix, respectively (Fig. 4F). C13 and C15 directly interact with protein side chains from the β2 and β3 strands, whereas A14 is the only base to be hydrogen bonded to amide and carbonyl of the protein main chain (Ile41) (Fig. 4F). This structure revealed that the NOVA2 KH3 domain interacts specifically with the 5′-UCAY-3′ RNA sequence. This information has been crucial for the in vivo identification of many new Nova binding sites and for a better understanding of the splicing regulation by this protein.70-72
Since only four bases can be accommodated by KH domains, we wondered how diverse were the sequences recognized by these RBDs. The available structures (Table 3) reveal that the motifs UCAC, UAAC, TCCC, CCCT, AGAA, CAAU, ATTC and TTTT were found interacting with NOVA KH3, SF1 KH, hnRNPK KH3, PCBP2 KH3, NusA KH1 and KH2, FBP KH3 and KH4, respectively. Surprisingly, although the mode of interaction of these domains with nucleic acids seems not to be as versatile as RRMs or ZnFs, KH domains can nevertheless bind a large panel of sequences. Comparing the RNA binding interactions reported for these three RNA binding domains, another difference is the absence of inter RNA-protein stacking interactions described for KH domains. This particularity could in part explain the low affinity (micromolar range) observed for these motifs interacting with single stranded nucleic acids. In order to counteract this apparent low specificity and affinity at least two strategies were selected during the evolution of these RBDs.
How KH Domains Increase Their Affinity and Specificity of Interaction
The first strategy consists in extending the KH domain surface of interaction with nucleic acids. The splicing factor SF1/mBBP is a good example since it specifically binds the 5′-UACUAAC-3′ intron branchpoint sequence (BPS) in human pre-mRNA transcripts73 using a binding surface composed of a KH domain extended by a C-terminal helix known as the QUA2 domain (Quaking homology 2)68 (Fig. 1). This extended KH surface with a βααββαα topology enables the binding of six nucleotides instead of the four nucleotides usually bound by a single KH domain. The 3′-end of the BPS (5′-UAAC-3′), which contains the conserved branch point adenosine (underlined), is specifically recognized by the KH domain, whereas the 5′-end (5′-ACU-3′) is bound by conserved residues from the QUA2 domain (Fig. 4G). In good agreement with the conservation of the branch point adenosine, the NMR structure of this complex shows that this base is specifically recognized by hydrogen bonds involving the main chain of Ile17768 similar to the contact to A14 in Nova2 KH3 with Ile41 (Fig. 4F). Another example of extension was also reported for the KH4 domain of the KSRP protein which contains a fourth β-strand located adjacent to the β1-strand (2HH2).74 However, this additional secondary element has still not been shown to be involved in binding nucleic acids.
The second feature consists in the repetition of multiple KH domains within a single RNA binding protein. As described for RRMs, these domains can either act independently or cooperatively. FUSE-binding protein (FBP) contains four KH domains and regulates c-myc expression by binding to FUSE.75 The structure of FBP KH domains 3 and 4 was solved by NMR in complex with a 29 nucleotide ssDNA molecule derived from its FUSE binding site. Each KH domain binds separately its DNA target. They behave independently without any contact between each other due to the presence of a glycine-rich flexible linker (30 amino acids) separating both KH domains. As observed for RRMs the presence of multiple RBDs in a single protein increases their chance to bind their targets especially when each domain binds nucleic acid molecules with a weak affinity. On the contrary, the two KH domains of NusA are separated by a short linker. It results in extensive contacts between the two domains forming a continuous platform of interaction for the targeted RNA. The 5′-end and 3′-end of the RNA interact within the cleft of KH1 and KH2, respectively binding together RNA with a nanomolar affinity.63 Similarly, the KH3 and KH4 domains of KSRP were also shown binding their ligand more tightly than each separated.74
In this section, we showed that KH domains are able to interact with a large panel of four-nucleotide long sequences. However, contrary to zinc fingers, they always keep the same fold and adapt their recognition mode by subtle variations of the atom sets involved in nucleic acid binding. Importantly, although a single KH domain binds nucleic acid molecules with a rather weak affinity, when present in multiple copies they can act in synergy and interact efficiently with their targets.
THE dsRBMs
dsRBMs Bind Double Stranded RNAs
Contrary to the three families of RNA binding domains described above, dsRBMs (double-stranded RNA binding motifs) were first described as recognizing RNA shape rather than RNA sequence.76 Typically, these domains contain approximately 70 amino acids and exhibit a conserved αβββα protein topology. They are often found in multiple copies (up to five in Drosophila melanogaster Staufen protein) and are involved in multiple functions such as RNP localization, RNA interference, RNA processing, RNA localisation, RNA editing and translational control.77 Until now, only few structures of dsRBMs in complex with RNA have been solved78-83 (Table 4).
Table 4.
Structures of dsRBMs solved in complex with RNA. The protein domains and the method used for the structure determination are indicated with the corresponding PDB number (http://www.rcsb.org/pdb/home/home.do) .
Based on these structures some common features can be observed about the mode of dsRBM interaction with RNA. These domains all interact along one face of a regular A-form helix structure and can cover up to 16 bp (e.g., Xlrbpa281) spanning two consecutive minor grooves separated by a major groove (Fig. 4H). In most of the cases, dsRBMs use residues from the α1 helix and β1-β2 loop (loop 2) to contact the minor grooves and N-terminus of the α2 helix with the preceding loop (loop 4) to bind the major groove (Fig. 4H). In addition, it was previously reported that the spacing between the loops 2 and 4 of dsRBMs fits better with the distance separating the minor and major grooves of RNA A-type helices than with the equivalent distance found in the B-helix form of dsDNA molecules.77 Based on this last observation and on the multiple interactions described between dsRBMs and 2′-OH groups of RNA riboses, these binding domains were first described as recognizing preferentially the double-stranded RNA shape.76,77
Interestingly, some particularities have also been emphasized with structures of dsRBMs in complex with RNA. In dsRBM of Xlrbpa2, the α1 helix interacts non specifically with the minor groove of the RNA via few contacts to the bases.81 These interactions were reported to be mostly mediated by water molecules. In dsRBM3 of Staufen, the α1 helix interacts with a UUCG tetraloop that caps the RNA double helix.80 In the dsRBM of Rnt1p, the α3 helix stabilizes the conformation of the α1 helix which contacts the sugar-phosphate backbone of the RNA minor groove and two nonconserved bases of the AGNN tetraloop.84 Finally, the structure of ADAR2 dsRBMs in complex with RNA has recently revealed that these domains were sometimes also able to bind sequence specifically dsRNAs82 (see below).
Some dsRBMs Interact Specifically with dsRNAs
ADAR2 is a human dsRBM containing protein that converts adenosine-to-inosine (A-to-I) by hydrolytic deamination in numerous mRNA and pre-mRNA transcripts.85,86 This protein has a modular domain organization consisting of two dsRBMs followed by a conserved C-terminal catalytic adenosine deaminase domain (Fig. 1). The structure of ADAR2 dsRBM1 and dsRBM2 was solved by NMR in complex with a stem-loop containing an A-to-I editing site.82 Both dsRBMs interact similarly with their targeted dsRNA. Lysine residues of a well conserved KKNAK motif located in the N-terminal part of the α2 helix interact nonspecifically with phosphate oxygens of residues from the major groove (Fig. 4H). More unexpected was the fact that each dsRBM binds the RNA stem-loop at a single register with sequence-specific contacts in the minor grooves. The amino group of a guanine is specifically recognized via a hydrogen bond formed with a main chain carbonyl of the β1-β2 loop and a hydrophobic contact is observed between a methionine side chain from the α1 helix and the proton at position 2 of an adenine (Fig. 4H). It was the first time that some dsRBMs were shown recognizing not only the shape of the RNA but also the sequence.82 More specifically, the structure explains the strong preference for a guanosine moiety 3′ to the edited adenosine since dsRBM2 of ADAR2 specifically recognizes the amino group of this base. In interacting with this nucleotide and the one which base-pairs with the editing site, dsRBM2 not only brings the deaminase domain in close proximity to the editing site, but also does not prevent access of the adenosine to the deaminase domain.82 Finally, this structural study explains how the edited adenine is targeted specifically by ADAR2 among the numerous other adenines located in the stem.
As for ADAR2, sequence specific contacts could also be observed between the α1 helix and β1-β2 loop of the Aa RNase III dsRBM and the RNA minor grooves.78,82 However, one difference is that dsRBM of Aa RNase III preferentially recognizes an RNA helix containing a G-X10-G sequence, whereas dsRBM1 and dsRBM2 of ADAR2 bind G-X9-A and G-X8-A, respectively. The length and the positioning of the α1 helix relative to the dsRBM fold appear to be the key structural elements that determine the register length of the different dsRBMs.82 Surprisingly, alignment of several dsRBM sequences reveals a high variability in the length and amino acid sequence composition of the α1 helix and the β1-β2 loop.82 In agreement with reports indicating that dsRBMs from different proteins are not functionally interchangeable, it strongly suggests that dsRBMs are likely to have different binding specificities.87,88
CONCLUSION AND PERSPECTIVES
In this chapter, we have described the current knowledge of how different RBDs interact with RNA at the atomic level and participate in RBP functions. Although still few structures of RBD containing proteins bound to RNA have been determined compared to the vast number of RNA binding proteins, few conclusions or hypotheses can be nevertheless drawn from these structures.
Importantly, the common determinant of the four RBD families described in this chapter is their ability to interact specifically with RNA. Structural biology highly contributed to provide crucial information about this specificity of interaction. For example, it was essential to correctly map binding sites for several splicing factors in vivo (the best examples are Fox-1 and NOVA2), since it revealed that the positioning of these binding sites relative to the splice-sites appears to be a major element controlling the mode of action of these proteins. Although this information is not sufficient to fully characterize this mode of action, it contributes to a better understanding of their functions. It also helped to understand how PTB, U2AF65 and Sex-lethal adapt to the different pyrimidine-tracts found at the 3′ splice-site and how Tra2-β1 recruits additional splicing factors on the SMN exon7. Finally, solving the structures of RBPs bound to RNA revealed unexpected features like the potential for RNA looping by PTB or MBNL1 suggesting a new function for these proteins in remodelling RNA structure.
However, it is still very hard to predict RBD-RNA interactions due to their versatility of interaction. We showed that the extreme plasticity of RRM for binding RNA can be explained by the use of different combinations of side chain and main chain RNA interactions but also by the capacity for this domain to increase its RNA binding surface outside the canonical β-sheet surface, using an additional β-strand, loops and/or RRM extremities. Zinc fingers use another strategy. Rather than extending their binding surface, they adopt different folds as emphasize in this chapter with the ZRANB2 family. On the contrary KH domains always use the same surface of interaction but still bind different sequences. This variability of RBD-RNA interactions justify the need to determine still more structures of protein-RNA complexes.
Despite progress in the last decade in this growing field, many questions remain to be answered. This ranges from simple questions that could be addressed rapidly by a structural biology approach to more complicated ones that will require multidisciplinary approaches or new methodologies. For example, we still need to address how pseudo-RRMs bind RNA. A more challenging question is how several RBPs assemble or multimerise on RNA? Also, how dynamic are protein-RNA interactions and how posttranslational modifications such as phosphorylation influence this dynamic? Answers to these questions are now needed for a full understanding of posttranscriptional gene regulations.
ACKNOWLEDGEMENTS
The authors would like to thank the Swiss National Science Foundation (No. 31003AB-133134), the SNF-NCCR Structural Biology and EURASNET for financial support to FHTA and the European Molecular Biology Organization for a postdoctoral fellowship to AC.
REFERENCES
- 1.
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. [PubMed: 11994740]
- 2.
- Lorsch JR. RNA chaperones exist and DEAD box proteins get a life. Cell. 2002;109:797–800. [PubMed: 12110176]
- 3.
- Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. [PubMed: 11181995]
- 4.
- Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate posttranscriptional gene expression. FEBS J. 2005;272:2118–2131. [PubMed: 15853797]
- 5.
- Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struc Biol. 2008;18:290–298. [PubMed: 18515081]
- 6.
- Oubridge C, Ito N, Evans PR, et al. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. [PubMed: 7984237]
- 7.
- Deo RC, Bonanno JB, Sonenberg N, et al. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. [PubMed: 10499800]
- 8.
- Handa N, Nureki O, Kurimoto K, et al. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. [PubMed: 10217141]
- 9.
- Hardin JW, Hu YX, McKay DB. Structure of the RNA binding domain of a DEAD-box helicase bound to its ribosomal RNA target reveals a novel mode of recognition by an RNA recognition motif. J Mol Biol. 2010;402:412–427. [PMC free article: PMC2942969] [PubMed: 20673833]
- 10.
- Kotik-Kogan O, Valentine ER, Sanfelice D, et al. Structural analysis reveals conformational plasticity in the recognition of RNA 3' ends by the human La protein. Structure. 2008;16:852–862. [PMC free article: PMC2430598] [PubMed: 18547518]
- 11.
- Lunde BM, Horner M, Meinhart A. Structural insights into cis element recognition of nonpolyadenylated RNAs by the Nab3-RRM. Nucleic Acids Research. 2011;39:337–346. [PMC free article: PMC3017603] [PubMed: 20805243]
- 12.
- Pancevac C, Goldstone DC, Ramos A, et al. Structure of the Rna15 RRM-RNA complex reveals the molecular basis of GU specificity in transcriptional 3'-end processing factors. Nucleic Acids Res. 2010;38:3119–3132. [PMC free article: PMC2875009] [PubMed: 20097654]
- 13.
- Pomeranz Krummel DA, Oubridge C, et al. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. [PMC free article: PMC2673513] [PubMed: 19325628]
- 14.
- Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B′′-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. [PubMed: 9716128]
- 15.
- Sickmier EA, Frato KE, Shen H, et al. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF 65. Mol Cell. 2006;23:49–59. [PMC free article: PMC2043114] [PubMed: 16818232]
- 16.
- Teplova M, Song J, Gaw HY, et al. Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure. 2010;18:1364–1377. [PMC free article: PMC3381513] [PubMed: 20947024]
- 17.
- Teplova M, Yuan YR, Phan AT, et al. Structural basis for recognition and sequestration of UUU(OH) 3' temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell. 2006;21:75–85. [PMC free article: PMC4689297] [PubMed: 16387655]
- 18.
- Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol. 2001;8:141–145. [PubMed: 11175903]
- 19.
- Yang Q, Coseno M, Gilmartin GM, et al. Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure. 2011;19:368–377. [PMC free article: PMC3056899] [PubMed: 21295486]
- 20.
- Allain FH, Bouvet P, Dieckmann T, et al. Molecular basis of sequence-specific recognition of preribosomal RNA by nucleolin. Embo J. 2000;19:6870–6881. [PMC free article: PMC305906] [PubMed: 11118222]
- 21.
- Allain FH, Howe PW, Neuhaus D, et al. Structural basis of the RNA-binding specificity of human U1A protein. Embo J. 1997;16:5764–5772. [PMC free article: PMC1170207] [PubMed: 9312034]
- 22.
- Auweter SD, Fasan R, Reymond L, et al. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. Embo J. 2006;25:163–173. [PMC free article: PMC1356361] [PubMed: 16362037]
- 23.
- Clery A, Jayne S, Benderska N, et al. Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1. Nat Struct Mol Biol. 2011;18:443–450. [PubMed: 21399644]
- 24.
- Dominguez C, Fisette JF, Chabot B, et al. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat Struct Mol Biol. 2010;17:853–861. [PubMed: 20526337]
- 25.
- Hargous Y, Hautbergue GM, Tintaru AM, et al. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. Embo J. 2006;25:5126–5137. [PMC free article: PMC1630407] [PubMed: 17036044]
- 26.
- Johansson C, Finger LD, Trantirek L, et al. Solution structure of the complex formed by the two N-terminal RNA-binding domains of nucleolin and a pre-rRNA target. J Mol Biol. 2004;337:799–816. [PubMed: 15033352]
- 27.
- Martin-Tumasz S, Reiter NJ, Brow DA, et al. Structure and functional implications of a complex containing a segment of U6 RNA bound by a domain of Prp24. RNA. 2010;16:792–804. [PMC free article: PMC2844626] [PubMed: 20181740]
- 28.
- Oberstrass FC, Auweter SD, Erat M, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. [PubMed: 16179478]
- 29.
- Perez-Canadillas JM. Grabbing the message: structural basis of mRNA 3'UTR recognition by Hrp1. Embo J. 2006;25:3167–3178. [PMC free article: PMC1500993] [PubMed: 16794580]
- 30.
- Skrisovska L, Bourgeois CF, Stefl R, et al. The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction. Embo Rep. 2007;8:372–379. [PMC free article: PMC1852761] [PubMed: 17318228]
- 31.
- Tsuda K, Kuwasako K, Takahashi M, et al. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res. 2009;37:5151–5166. [PMC free article: PMC2731918] [PubMed: 19553194]
- 32.
- Tsuda K, Someya T, Kuwasako K, et al. Structural basis for the dual RNA-recognition modes of human Tra2-beta RRM. Nucleic Acids Res. 2011;39:1538–1553. [PMC free article: PMC3045587] [PubMed: 20926394]
- 33.
- Varani L, Gunderson SI, Mattaj IW, et al. The NMR structure of the 38 kDa U1A protein—PIE RNA complex reveals the basis of co-operativity in regulation of polyadenylation by human U1A protein. Nat Struct Biol. 2000;7:329–335. [PubMed: 10742179]
- 34.
- Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. [PMC free article: PMC1635273] [PubMed: 16982642]
- 35.
- Allain FH, Gubser CC, Howe PW, et al. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formulation. Nature. 1996;380:646–650. [PubMed: 8602269]
- 36.
- Jacks A, Babon J, Kelly G, et al. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure. 2003;11:833–843. [PubMed: 12842046]
- 37.
- Avis JM, Allain FH, Howe PW, et al. Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding. J Mol Biol. 1996;257:398–411. [PubMed: 8609632]
- 38.
- Perez Canadillas JM, Varani G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. Embo J. 2003;22:2821–2830. [PMC free article: PMC156756] [PubMed: 12773396]
- 39.
- Mazza C, Segref A, Mattaj IW, et al. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. Embo J. 2002;21:5548–5557. [PMC free article: PMC129070] [PubMed: 12374755]
- 40.
- Hofmann Y, Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum Mol Genet. 2002;11:2037–2049. [PubMed: 12165565]
- 41.
- Young PJ, DiDonato CJ, Hu D, et al. SRp30c-dependent stimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2 beta 1. Hum Mol Genet. 2002;11:577–587. [PubMed: 11875052]
- 42.
- Dominguez C, Allain FH. NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: a novel mode of RNA recognition. Nucleic Acids Res. 2006;34:3634–3645. [PMC free article: PMC1540728] [PubMed: 16885237]
- 43.
- Blanchette M, Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. Embo J. 1999;18:1939–1952. [PMC free article: PMC1171279] [PubMed: 10202157]
- 44.
- Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. [PubMed: 7761834]
- 45.
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Bioph Biom. 2000;29:183–212. [PubMed: 10940247]
- 46.
- Amarasinghe GK, De Guzman RN, Turner RB, et al. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J Mol Biol. 2000;301:491–511. [PubMed: 10926523]
- 47.
- De Guzman RN, Wu ZR, Stalling CC, et al. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. [PubMed: 9430589]
- 48.
- Dey A, York D, Smalls-Mantey A, et al. Composition and sequence-dependent binding of RNA to the nucleocapsid protein of Moloney murine leukemia virus. Biochemistry-US. 2005;44:3735–3744. [PubMed: 15751950]
- 49.
- Zhou J, Bean RL, Vogt VM, et al. Solution structure of the Rous sarcoma virus nucleocapsid protein: muPsi RNA packaging signal complex. J Mol Biol. 2007;365:453–467. [PMC free article: PMC1764217] [PubMed: 17070546]
- 50.
- Hudson BP, Martinez-Yamout MA, Dyson HJ, et al. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–264. [PubMed: 14981510]
- 51.
- Lee BM, Xu J, Clarkson BK, et al. Induced fit and «lock and key» recognition of 5S RNA by zinc fingers of transcription factor IIIA. J Mol Biol. 2006;357:275–291. [PubMed: 16405997]
- 52.
- Loughlin FE, Mansfield RE, Vaz PM, et al. The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5' splice site-like sequences. Proc Nat Acad Sci USA. 2009;106:5581–5586. [PMC free article: PMC2667063] [PubMed: 19304800]
- 53.
- Lu D, Searles MA, Klug A. Crystal structure of a zinc-finger-RNA complex reveals two modes of molecular recognition. Nature. 2003;426:96–100. [PubMed: 14603324]
- 54.
- Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol. 2008;15:1343–1351. [PMC free article: PMC4689322] [PubMed: 19043415]
- 55.
- Engelke DR, Ng SY, Shastry BS, et al. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980;19:717–728. [PubMed: 6153931]
- 56.
- Pelham HR, Brown DD. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proceedings of the National Academy of Sciences of the United States of America. 1980;77:4170–4174. [PMC free article: PMC349792] [PubMed: 7001457]
- 57.
- Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc T. 2002;30:945–952. [PubMed: 12440952]
- 58.
- Pascual M, Vicente M, Monferrer L, et al. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. [PubMed: 16533306]
- 59.
- Plambeck CA, Kwan AH, Adams DJ, et al. The structure of the zinc finger domain from human splicing factor ZNF265 fold. J Biol Chem. 2003;278:22805–22811. [PubMed: 12657633]
- 60.
- Nguyen CD, Mansfield RE, Leung W, et al. Characterization of a family of RanBP2-type zinc fingers that can recognize single-stranded RNA. J Mol Biol. 2011;407:273–283. [PubMed: 21256132]
- 61.
- Grishin NV. KH domain: one motif, two folds. Nucleic Acids Res. 2001;29:638–643. [PMC free article: PMC30387] [PubMed: 11160884]
- 62.
- Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275:2712–2726. [PubMed: 18422648]
- 63.
- Beuth B, Pennell S, Arnvig KB, et al. Structure of a Mycobacterium tuberculosis NusA-RNA complex. Embo J. 2005;24:3576–3587. [PMC free article: PMC1276712] [PubMed: 16193062]
- 64.
- Braddock DT, Louis JM, Baber JL, et al. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature. 2002;415:1051–1056. [PubMed: 11875576]
- 65.
- Du Z, Lee JK, Fenn S, et al. X-ray crystallographic and NMR studies of protein-protein and protein-nucleic acid interactions involving the KH domains from human poly(C)-binding protein-2. RNA. 2007;13:1043–1051. [PMC free article: PMC1894928] [PubMed: 17526645]
- 66.
- Jia MZ, Horita S, Nagata K, et al. An archaeal Dim2-like protein, aDim2p, forms a ternary complex with a/eIF2 alpha and the 3' end fragment of 16S rRNA. J Mol Biol. 2010;398:774–785. [PubMed: 20363226]
- 67.
- Lewis HA, Musunuru K, Jensen KB, et al. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. [PubMed: 10676814]
- 68.
- Liu Z, Luyten I, Bottomley MJ, et al. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science. 2001;294:1098–1102. [PubMed: 11691992]
- 69.
- Tu C, Zhou X, Tropea JE, et al. Structure of ERA in complex with the 3' end of 16S rRNA: implications for ribosome biogenesis. Proc Nat Acad Sci USA. 2009;106:14843–14848. [PMC free article: PMC2736428] [PubMed: 19706445]
- 70.
- Ule J, Ule A, Spencer J, et al. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005;37:844–852. [PubMed: 16041372]
- 71.
- Ule J, Jensen KB, Ruggiu M, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. [PubMed: 14615540]
- 72.
- Ule J, Stefani G, Mele A, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. [PubMed: 17065982]
- 73.
- Berglund JA, Chua K, Abovich N, et al. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. [PubMed: 9182766]
- 74.
- Garcia-Mayoral MF, Hollingworth D, Masino L, et al. The structure of the C-terminal KH domains of KSRP reveals a noncanonical motif important for mRNA degradation. Structure. 2007;15:485–498. [PubMed: 17437720]
- 75.
- Michelotti GA, Michelotti EF, Pullner A, et al. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol Cell Biol. 1996;16:2656–2669. [PMC free article: PMC231256] [PubMed: 8649373]
- 76.
- Stefl R, Skrisovska L, Allain FH. RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle. Embo Rep. 2005;6:33–38. [PMC free article: PMC1299235] [PubMed: 15643449]
- 77.
- Chang KY, Ramos A. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 2005;272:2109–2117. [PubMed: 15853796]
- 78.
- Gan J, Tropea JE, Austin BP, et al. Structural insight into the mechanism of double-stranded RNA processing by ribonuclease III. Cell. 2006;124:355–366. [PubMed: 16439209]
- 79.
- Leulliot N, Quevillon-Cheruel S, Graille M, et al. A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III. Embo J. 2004;23:2468–2477. [PMC free article: PMC449770] [PubMed: 15192703]
- 80.
- Ramos A, Grunert S, Adams J, et al. RNA recognition by a Staufen double-stranded RNA-binding domain. Embo J. 2000;19:997–1009. [PMC free article: PMC305639] [PubMed: 10698941]
- 81.
- Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. Embo J. 1998;17:7505–7513. [PMC free article: PMC1171094] [PubMed: 9857205]
- 82.
- Stefl R, Oberstrass FC, Hood JL, et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–237. [PMC free article: PMC2956598] [PubMed: 20946981]
- 83.
- Yang SW, Chen HY, Yang J, et al. Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure. 2010;18:594–605. [PMC free article: PMC3119452] [PubMed: 20462493]
- 84.
- Wu H, Henras A, Chanfreau G, et al. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc Nat Acad Sci USA. 2004;101:8307–8312. [PMC free article: PMC420390] [PubMed: 15150409]
- 85.
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. [PMC free article: PMC1823043] [PubMed: 12045112]
- 86.
- Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7:919–931. [PMC free article: PMC2953463] [PubMed: 17139332]
- 87.
- Liu Y, Lei M, Samuel CE. Chimeric double-stranded RNA-specific adenosine deaminase ADAR1 proteins reveal functional selectivity of double-stranded RNA-binding domains from ADAR1 and protein kinase PKR. Proc Nat Acad Sci USA. 2000;97:12541–12546. [PMC free article: PMC18800] [PubMed: 11070079]
- 88.
- Parker GS, Maity TS, Bass BL. dsRNA binding properties of RDE-4 and TRBP reflect their distinct roles in RNAi. J Mol Biol. 2008;384:967–979. [PMC free article: PMC2605707] [PubMed: 18948111]
- 89.
- Ding J, Hayashi MK, Zhang Y, et al. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Gene Dev. 1999;13:1102–1115. [PMC free article: PMC316951] [PubMed: 10323862]
- 90.
- Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14(51-55):29–32. [PubMed: 8744573]
- FROM STRUCTURE TO FUNCTION OF RNA BINDING DOMAINS - Madame Curie Bioscience Data...FROM STRUCTURE TO FUNCTION OF RNA BINDING DOMAINS - Madame Curie Bioscience Database
- Regulation and Coordination of Intracellular Trafficking: An Overview - Madame C...Regulation and Coordination of Intracellular Trafficking: An Overview - Madame Curie Bioscience Database
- Pre-Ribosomal RNA Processing in Multicellular Organisms - Madame Curie Bioscienc...Pre-Ribosomal RNA Processing in Multicellular Organisms - Madame Curie Bioscience Database
- Regulation of T Cell Immunity by OX40 and OX40L - Madame Curie Bioscience Databa...Regulation of T Cell Immunity by OX40 and OX40L - Madame Curie Bioscience Database
- Overview Αβ Metabolism: From Alzheimer Research to Brain Aging Control - Madame ...Overview Αβ Metabolism: From Alzheimer Research to Brain Aging Control - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...