All Medical Genetics Summaries content, except where otherwise noted, is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license which permits copying, distribution, and adaptation of the work, provided the original work is properly cited and any changes from the original work are properly indicated. Any altered, transformed, or adapted form of the work may only be distributed under the same or similar license to this one.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Pratt VM, Scott SA, Pirmohamed M, et al., editors. Medical Genetics Summaries [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2012-.
Introduction
Phenytoin (brand name Dilantin) is an anticonvulsant medication used for the treatment of seizures (1).
Phenytoin has a narrow therapeutic index—individuals that have supratherapeutic blood concentrations of phenytoin have increased risks of acute side effects. Dosing can be complex due to pharmacokinetic factors, including individual weight, age, gender, concomitant medications, plasma binding protein status, the presence of uremia or hyperbilirubinemia, and specific pharmacogenetic variants. As such, therapeutic drug monitoring is often used to adjust dose and maintain serum concentrations within the therapeutic range (10–20 μg/mL).
The CYP2C9 enzyme is one of the main enzymes involved in the metabolism of phenytoin, and variant CYP2C9 alleles are known to influence phenytoin drug levels. Individuals who have decreased activity CYP2C9 variants may have reduced clearance rates of phenytoin and be at greater risk for dose-related side effects (2).
An individual’s human leukocyte antigen B (HLA-B) genotype is a known risk factor for drug-induced hypersensitivity reactions. The HLA-B protein has an important immunological role in pathogen recognition and response, as well as to non-pathogens such as drugs. Individuals who have the HLA-B*15:02 allele are at high risk of developing potentially life-threatening phenytoin-induced Stevens-Johnson syndrome (SJS) and the related toxic epidermal necrolysis (TEN).
The HLA-B*15:02 allele is most often found among individuals of Southeast Asian descent, where there is a strong association between SJS/TEN and exposure to carbamazepine. Carbamazepine is an antiseizure medication used to treat the same types of seizures as phenytoin, as well as trigeminal neuralgia and bipolar disorder.
The FDA-approved drug label for phenytoin states that consideration should be given to avoiding phenytoin as an alternative for carbamazepine in individuals positive for HLA-B*15:02 (Table 1). The label also mentions that variant CYP2C9 alleles may contribute to unusually high levels of phenytoin (1).
Dosing recommendations for phenytoin based on HLA-B and CYP2C9 genotype have also been published by the Clinical Pharmacogenetics Implementation Consortium (CPIC, Table 2, Figure 1) and the Dutch Pharmacogenetics Working Group (DPWG, Table 3, Table 4). These recommendations include the use of an antiseizure medication other than carbamazepine, phenytoin (or its prodrug fosphenytoin) for any HLA-B*15:02 positive individual regardless of CYP2C9 genotype, individual ancestry, or age. These recommendations also include specific dose reductions of phenytoin for individuals who have low or deficient enzyme activity (2, 3).
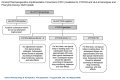
Figure 1:
Dosage Guidelines from the CPIC for Phenytoin based on HLA-B and CYP2C9 Genotype. Figure reproduced with permission from the authors.
Table 1.
FDA Phenytoin Dosage and Administration (2019)
| Gene or gene variant | Dosing considerations |
|---|---|
| HLA-B*15:02 | Consideration should be given to avoiding phenytoin as an alternative for carbamazepine in individuals positive for HLA-B*15:02. The use of HLA-B*15:02 genotyping has important limitations and must never substitute for appropriate clinical vigilance and individual management. |
| CYP2C9 and CYP2C19 | If individual is phenytoin naïve, do not use phenytoin/fosphenytoin. Avoid carbamazepine and oxcarbazepine.a If the individual has previously used phenytoin continuously for longer than 3 months without incidence of cutaneous adverse reactions, cautiously consider use of phenytoin in the future. The latency period for drug-induced SJS/TEN is short with continuous dosing and adherence to therapy (4–28 days), and cases usually occur within 3 months of dosing.b |
This FDA table is adapted from (1).
Table 2.
The CPIC Recommended Dosing of Phenytoin Based on HLA-B*15:02 and CYP2C9 Phenotype/Genotype (2020)
| CYP2C9 phenotype and HLA-B genotype | Implication | Therapeutic recommendation |
|---|---|---|
| Any CYP2C9 phenotype and HLA-B*15:02 positive# | Increased risk of phenytoin-induced SJS/ TEN | If the individual is phenytoin-naïve, do not use phenytoin/fosphenytoin. Avoid carbamazepine and oxcarbazepine.a If the individual has previously used phenytoin continuously for longer than 3 months without incidence of cutaneous adverse reactions, cautiously consider use of phenytoin in the future. The latency period for drug-induced SJS/TEN is short with continuous dosing and adherence to therapy (428 days), and cases usually occur within 3 months of dosing.b |
| CYP2C9 normal metabolizer and HLA-B*15:02 negative | Normal phenytoin metabolism | No adjustments needed from typical dosing strategies. Subsequent doses should be adjusted according to therapeutic drug monitoring, response, and side effects. An HLA-B*15:02 negative test does not eliminate the risk of phenytoin-induced SJS/TEN, and individuals should be carefully monitored according to standard practice.a |
| CYP2C9 intermediate metabolizer (activity score 1.5) and HLA-B*15:02 negative | Slightly reduced phenytoin metabolism: however, this does not appear to translate into increased side effects. | No adjustments needed from typical dosing strategies. Subsequent doses should be adjusted according to therapeutic drug monitoring, response, and side effects. An HLA-B*15:02 negative test does not eliminate the risk of phenytoin-induced SJS/TEN, and individuals should be carefully monitored according to standard practice.c |
| CYP2C9 intermediate metabolizer (Activity score 1.0) and HLA-B*15:02 negative | Reduced phenytoin metabolism: Higher plasma concentrations will increase probability of toxicities. | For first dose, use typical initial or loading dose. For subsequent doses, use approximately 25% less than typical maintenance dose. Subsequent doses should be adjusted according to therapeutic drug monitoring, response, and side effects. An HLA-B*15:02 negative test does not eliminate the risk of phenytoin-induced SJS/TEN, and individuals should be carefully monitored according to standard practice.c |
| CYP2C9 poor metabolizer and HLA-B*15:02 negative | Reduced phenytoin metabolism: Higher plasma concentrations will increase probability of toxicities. | For first dose, use typical initial or loading dose. For subsequent doses use approximately 50% less than typical maintenance dose. Subsequent doses should be adjusted according to therapeutic drug monitoring, response, and side effects. An HLA-B*15:02 negative test does not eliminate the risk of phenytoin-induced SJS/TEN, and individuals should be carefully monitored according to standard practice.a |
SJS/TEN: Stevens-Johnson syndrome/toxic epidermal necrolysis.
- #
Considerations: Other aromatic anticonvulsants, including eslicarbazepine, lamotrigine, and phenobarbital, have weaker evidence linking SJS/TEN with the HLA-B*15:02 allele; however, caution should still be used in choosing an alternative agent. Previous tolerance of phenytoin is not indicative of tolerance to other aromatic anticonvulsants.
a The strength of the therapeutic recommendation is classified as “strong”.
b The strength of the therapeutic recommendation is classified as “optional”.
c The strength of the therapeutic recommendation is classified as “moderate”.
This CPIC table is adapted from (2).
Note: The nomenclature used in this table reflects the standardized pharmacogenetic terms proposed by CPIC (4).
Table 3.
The DPWG Phenytoin Dosing based on HLA-B*15:02 Genotype (2017)
| Genotype | Implication | Dosing recommendations |
|---|---|---|
| Positive for HLA-B*15:02 | Increased risk of the life-threatening cutaneous side effect Stevens-Johnson syndrome/toxic epidermal necrolysis (SJS/TEN). The calculated risk of phenytoin-induced SJS/TEN in individuals with HLA-B*15:02 is 0.65%. | Carefully weigh the risk of SJS/TEN against the benefits Avoid phenytoin if an alternative is possible
|
This DPWG table is adapted from (3).
Table 4.
The DPWG Phenytoin Dosing based on CYP2C9 Genotype (2018)
| Metabolizer type | Genotype | Side effects | Recommendations | ||
|---|---|---|---|---|---|
| Loading dose | Other doses | Advise the individual | |||
| CYP2C9 IM | *1/*2 | Genetic variation reduces conversion of phenytoin to inactive metabolites. This increases the risk of side effects. For the genotype group *1/*3+*3/*3, an increased risk of the life-threatening cutaneous side effects Stevens-Johnson Syndrome and toxic epidermal necrolysis has been observed, especially in Asians. | The loading dose does not need to be adjusted. | Use 75% of the standard dose* | Advise the individual to report side effects (such as ataxia, nystagmus, slurred speech, sedation or, especially in Asian individuals, rash) occur. |
| *1/*3 | |||||
| other | |||||
| CYP2C9 PM | *2/*2 | Use 50% of the standard dose* | |||
| *2/*3 | Use 50% of the standard dose* | ||||
| *3/*3 | Use 40% of the standard dose* | ||||
| other | Use 40–50% of the standard dose* | ||||
- *
Assess the dose based on effect and serum concentration after 7–10 days.
This DPWG table is adapted from (5).
IM – intermediate metabolizer
PM – poor metabolizer
Drug: Phenytoin
Phenytoin is a generic antiseizure drug that is rarely prescribed to newly diagnosed individuals due to its propensity for long-term side effects. Nevertheless, it continues to be used by many individuals who initiated treatment before the availability of newer medications that have fewer side effects and drug-drug interactions. Phenytoin is used for the control of partial seizures and generalized tonic-clonic convulsions. It is also used in the treatment of status epilepticus and may be used to prevent or treat seizures that occur during and following neurosurgery (1).
Phenytoin belongs to the sodium channel blocker class of antiseizure drugs, which are thought to suppress seizure activity by blocking voltage-gated sodium channels that are responsible for the upstroke of action potentials (6, 7). The block by phenytoin and other members of this class of antiseizure drugs occurs in a state-dependent fashion, with preferential binding and block of the inactivated state of the channel. This results in voltage- and frequency-dependent block in which high frequency action potential firing, which occurs during epileptic activity, is preferentially inhibited. (1, 8)
The dosing of phenytoin can be complex, as treatment is typically initiated at a low starting dose, which considers individual age, weight, and the presence of concomitant medications that may influence phenytoin metabolism or protein binding. The dose is then carefully escalated to obtain the desired therapeutic effect. There is a wide variation in how individuals respond to phenytoin (2). Therapeutic drug monitoring is often used to adjust the dose to ensure that plasma levels are within therapeutic range (10–20 μg/ml in adults). Measurement of plasma levels is useful when adding or discontinuing concomitant medications that effect phenytoin levels. Periodic measurement of plasma phenytoin concentrations may also be valuable in pregnancy because altered phenytoin pharmacokinetics increases the risk of seizures.
Phenytoin use during pregnancy has been associated with an 11% risk of fetal hydantoin syndrome in the offspring, which is characterized by dysmorphism, hypoplasia, and irregular ossification of the distal phalanges. Facial dysmorphism includes epicanthal folds, hypertelorism, broad flat nasal bridges, an upturned nasal tip, wide prominent lips, and, in addition, distal digital hypoplasia, intrauterine growth retardation, and mental retardation. An additional 30% of the in utero-exposed children express fetal hydantoin effects, in which there is a more limited pattern of dysmorphic characteristics. Some studies have found significant associations between in utero exposure to phenytoin and major congenital abnormalities (mainly, cardiac malformations and cleft palate) whereas others have failed to find such associations (9, 10).
The adverse effects of phenytoin fall into 2 categories, types A and B. (11)
Type A adverse drug reactions account for up to 90% of reactions. They are predictable and can occur in any individual if their drug exposure is high enough. Some of these reactions occur rapidly and are reversible when the dose is reduced. These include acute central nervous system adverse effects such as sedation, nystagmus, and ataxia. Other common side effects occur with long-term exposure and include changes to the physical appearance, such as gingival hyperplasia, coarsening of the facial features, hirsutism, and acne.
Type B adverse drug reactions include idiosyncratic hypersensitivity reactions, such as severe cutaneous adverse reaction (SCAR). Such reactions can occur at any dose and develop through a mechanism that is unrelated to the mechanism of action of the drug. A rare but life-threatening hypersensitivity reaction associated with phenytoin treatment is SJS and the related TEN. Both are severe cutaneous reactions to specific drugs, and are characterized by fever and lesions of the skin and mucous membranes, with a mortality rate of up to 30% (12).
It is difficult to predict in whom a drug-induced hypersensitivity reaction is likely to occur. However, for phenytoin individuals who are positive for a specific HLA allele are known to be susceptible to phenytoin-induced SJS/TEN. Human leukocyte antigen testing of individuals can identify at-risk individuals so that an alternative drug can be used.
The HLA Gene Family
The human leukocyte antigen (HLA) genes are members of the major histocompatibility complex (MHC) gene family, which includes more than 200 genes. The MHC family has been subdivided into 3 subgroups based on the structure and function of the encoded proteins: Class I, Class II, and Class III. The class I region contains the genes encoding the HLA molecules HLA-A, HLA-B, and HLA-C. These molecules are expressed on the surfaces of almost all cells and play an important role in processing and presenting antigens. The class I gene region also contains a variety of other genes, many of which are not known to be involved in immune function.
An important role of HLA class I molecules is to present peptide fragments to immune cells (CD8+ T cells). Most of these peptides originate from the breakdown of normal cellular proteins (“self”). However, if foreign peptide fragments are presented, for example, from a pathogen, CD8+T cells will recognize the peptides as “non-self” and will be activated to release inflammatory cytokines and launch an immune response to dispose of the pathogen (or foreign body).
Because HLA molecules need to present such a wide variety of “self” and “non-self” peptides, the HLA genes are both numerous and highly polymorphic. More than 1,500 HLA-B alleles have been identified (13). The HLA allele nomenclature includes the HLA prefix, followed by the gene, an asterisk and a 2 digit number that corresponds to antigen specificity, and the assigned allele number (14). For example, the HLA-B*15:02 allele is composed of:
- HLA: the HLA prefix (the HLA region on chromosome 6)
- B: the B gene (a particular HLA gene in this region)
- 15: the allele group (historically determined by serotyping, namely, a group of alleles that share the same serotype)
- 02: the specific HLA allele (a specific protein sequence; determined by genetic analysis).
Additional digits have recently been added to the nomenclature to discriminate alleles that do not differ in the protein amino acid sequence, but differ in their genetic sequence (namely, due to synonymous and noncoding genetic variants).
Variation in HLA genes plays an important role in the susceptibility to autoimmune disease and infections, and they are also critical in the context of transplant surgery where better outcomes are observed if the donor and recipient are HLA-compatible.
More recently, HLA alleles have been associated with susceptibility to Type B adverse drug reactions. For example, HLA-B alleles have been associated with severe hypersensitivity reactions to abacavir (used to treat HIV), allopurinol (used to treat gout), and the antiepileptic drugs carbamazepine and phenytoin.
Gene: HLA-B*15:02
Individuals who have one or 2 copies of the high-risk HLA-B*15:02 allele are known as HLA-B*15:02 positive (Table 5).
Table 5.
The CPIC Assignment of Likely HLA-B Phenotype Based on Genotype (2020)
| HLA phenotype | Genotype | Examples of diplotypes |
|---|---|---|
| HLA-B*15:02 negative | Homozygous for any allele(s) other than HLA-B*15:02 | *X/*X a |
| HLA-B*15:02 positive | Homozygous or heterozygous variant | *15:02/*X a , *15:02/*15:02 |
The association between the HLA-B*15:02 allele and SJS/TEN was first reported with the use of carbamazepine in the Han Chinese population. In the initial study, all individuals who had carbamazepine-induced SJS/TEN were found to have HLA-B*15:02 (44/44, 100%), whereas the allele was much less common among carbamazepine-tolerant individuals (3/101, 3%)(15). In subsequent studies, this association was replicated, with an HLA-B*15:02 positivity frequency of 70–100% among cases of carbamazepine-induced SJS/TEN (16).
The HLA-B*15:02 allele was later associated with phenytoin-induced hypersensitivity reactions, including phenytoin-induced SJS in a Thai population and phenytoin-induced SJS/TEN in Chinese Asians (17, 18).
There are fewer studies on phenytoin-induced hypersensitivity than carbamazepine, and the strength of association between phenytoin and SJS/TEN is weaker than that of carbamazepine and SJS/TEN. However, from the evidence available, the FDA recommends consideration of avoiding phenytoin as an alternative treatment to carbamazepine in individuals who have HLA-B*15:02 (2).
The prevalence of carbamazepine-induced SJS/TEN is higher in populations where HLA-B*15:02 is more common. Of note, the HLA-B*15:02 allele frequency is highest in Southeast Asia, as populations from Hong Kong, Thailand, Malaysia, Vietnam, and parts of the Philippines have an allele frequency >15%. It is slightly lower (~10–13%) in Taiwan and Singapore, and around 4% in North China. South Asians, including Indians, appear to have an HLA-B*15:02 allele frequency of ~2–4%, with higher frequencies in some subpopulations. (15, 16, 17, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30)
The HLA-B*15:02 allele is rare (<1%) in East Asia (Japan and Korea) and among individuals who are not of Asian descent. For example, the allele is rare in Europeans, Hispanics, Africans, African Americans, and Native Americans. (16, 21)
Gene: CYP2C9
The cytochrome P450 superfamily (CYP450) is a large and diverse group of hepatic enzymes that form the major system for metabolizing lipids, hormones, toxins, and drugs. The CYP450 genes are very polymorphic and can result in reduced, absent, or increased enzyme activity.
The CYP2C9 enzyme metabolizes approximately 15% of clinically used drugs, and atypical metabolic activity caused by genetic variants in the CYP2C9 gene can play a major role in adverse drug reactions (31, 32).
The CYP2C9 gene is polymorphic, with more than 50 known alleles. Variation in CYP2C9 is thought to contribute to the pharmacogenetic variability in phenytoin metabolism. CYP2C9*1 is considered the wild-type allele when no variants are detected and is categorized as normal enzyme activity (2). Individuals who have 2 normal-function alleles (for example, CYP2C9 *1/*1) are classified as “normal metabolizers” (Table 6).
Table 6.
The CPIC Assignment of Likely CYP2C9 Phenotype Based on Genotype (2020)
| CYP2C9 phenotypea,b | Activity score | Genotype | Examples of diplotypes |
|---|---|---|---|
| Normal metabolizer | 2 | An individual with 2 normal-function alleles | *1/*1 |
| Intermediate metabolizer | 1.5 1 | An individual with one normal-function allele plus one decreased-function allele OR one normal-function allele plus one no-function allele OR 2 decreased-function alleles | *1/*2 *1/*3 *2/*2 |
| Poor metabolizer | 0.5 0 | An individual with one no-function allele plus one decreased-function allele; OR 2 no-function alleles | *2/*3 *3/*3 |
- a
Assignment of allele function and associated citations can be found at the CPIC website, also see (2).
- b
See the CYP2C9 Frequency Table in refs. 3 and 4 from (2) for population-specific allele and phenotype frequencies.
Note: There are no known cases of CYP2C9 ultrarapid metabolizers
This CPIC table has been adapted from (2).
For individuals who are CYP2C9 normal metabolizers, the recommended starting maintenance dose of phenytoin does not need to be adjusted based on genotype (2).
Two common allelic variants associated with reduced enzyme activity are CYP2C9*2 (p.Arg144Cys) and CYP2C9*3 (p.Ile359Leu). The *2 allele is more common in Caucasian (10–20%), than Asian (1–3%) or African (0–6%) populations. The *3 allele is less common (<10% in most populations) and is extremely rare in African populations. In African Americans, the CYP2C9*5, *6, *8 and *11 alleles are more common (33, 34, 35).
Linking HLA-B and CYP2C9 Genetic Variation with the Risk of Side Effects and Treatment Response
Reduced activity CYP2C9 alleles, in particular CYP2C9*3, influence phenytoin dosage, individual response, and predict adverse drug reactions (36, 37, 38, 39, 40). Individuals with reduced-function CYP2C9 alleles have reduced clearance of phenytoin and have an increased risk of side effects.
Specific HLA alleles, namely, HLA-B*15:02, are also strongly associated with SCAR (41).
To guide the optimal dose of phenytoin and reduce the risk, both genetic factors (CYP2C9 and HLA alleles) and non-genetic factors (for example, omeprazole co-medication) need to be considered (41, 42, 43, 44, 45).
The HLA-B and CYP2C9 Gene Interactions with Medications Used for Additional Indications
Other medications with multiple indications are known to interact with the HLA-B alleles or to be metabolized by CYP2C9.
- Other seizure medications—Carbamazepine has similar interactions with HLA variation and hypersensitivity reactions: individuals with one or more copies of HLA-B*15:02 are at risk of SJS/TEN and the HLA-A*31:01 is strongly associated with a potentially life-threatening condition known as drug reaction with eosinophilia and systemic symptoms (DRESS) and a milder reaction maculopapular exanthema (MPE).
- Uric acid reduction medications—Allopurinol is a xanthine oxidase inhibitor to treat high uric acid levels seen in gout, tumor lysis syndrome, and cases of symptomatic hyperuricemia. The HLA-B*58:01 allele is associated with SCAR during allopurinol treatment. Lesinurad is a urate transport inhibitor, also used in the treatment of gout and it is metabolized by CYP2C9 to inactive metabolites. Individuals who are CYP2C9 poor metabolizers will have an increased exposure to the active drug and thus have an increased risk of side effects such as kidney stones or cardiovascular events.
- Anti-retroviral medication—Abacavir, a nucleoside/nucleotide reverse transcriptase inhibitor used in the treatment of HIV, is also associated with hypersensitivity reactions, the risk of which increases when an individual has the HLA-B*57:01 allele.
- Non-steroidal anti-inflammatory drugs (NSAIDs)—Celecoxib, Flurbiprofen, and Piroxicam are used for pain management in osteoarthritis, rheumatoid arthritis, and other conditions; they are all metabolized by CYP2C9 and poor metabolizers will experience higher levels of exposure to these NSAIDs and have higher risk of side effects.
- Anti-emetics—Dronabinol, a synthetic cannabinoid, is used in the treatment of chemotherapy-induced nausea and vomiting for individuals who had poor responses to traditional anti-emetics. It also is used to treat anorexia-associated weight loss in individuals with acquired immunodeficiency syndrome. Dronabinol is activated by CYP2C9 metabolism and individuals with poor metabolizer phenotypes will have an increased exposure to dronabinol and increased risk of side effects.
Additional information on gene-drug interactions for HLA-B and CYP2C9 are available from PharmGKB, CPIC, and the FDA (search for “HLA-B” or “CYP2C9”).
Genetic Testing
The NIH’s Genetic Testing Registry provides examples of the genetic tests that are available for the phenytoin drug response, the HLA-B gene, and the CYP2C9 gene.
The genotype results for an HLA allele such as HLA-B*15:02 can either be “positive” or “negative”. There are no intermediate phenotypes because the HLA genes are expressed in a codominant manner.
A positive result indicates the individual is either “heterozygous” or “homozygous” for the variant, depending upon whether they have one or 2 copies of the *15:02 allele.
A negative result indicates that the individual does not have the HLA-B*15:02 allele. However, a negative result does not rule out the possibility of an individual developing phenytoin-induced SJS/TEN. Therefore, clinicians should carefully monitor all individuals according to standard practices.
For CYP2C9, alleles to be included in clinical genotyping assays have been recommended by the Association for Molecular Pathology. (46) Results are typically reported as a diplotype, such as CYP2C9 *1/*2, and may include an interpretation of the individual’s predicted metabolizer phenotype (normal, intermediate, or poor) and an activity score (Table 6).
Therapeutic Recommendations based on Genotype
This section contains excerpted1 information on gene-based dosing recommendations. Neither this section nor other parts of this review contain the complete recommendations from the sources.
2019 Statement from the US Food and Drug Administration (FDA)
Regarding HLA-B:
Studies in individuals of Chinese ancestry have found a strong association between the risk of developing SJS/TEN and the presence of HLA-B*15:02, an inherited allelic variant of the HLA-B gene, in individuals using carbamazepine. Limited evidence suggests that HLA-B*15:02 may be a risk factor for the development of SJS/TEN in individuals of Asian ancestry taking other antiepileptic drugs associated with SJS/TEN, including phenytoin. Consideration should be given to avoiding phenytoin as an alternative for carbamazepine in individuals positive for HLA-B*15:02.
The use of HLA-B*15:02 genotyping has important limitations and must never substitute for appropriate clinical vigilance and individual management. The role of other possible factors in the development of, and morbidity from, SJS/TEN, such as antiepileptic drug (AED) dose, compliance, concomitant medications, comorbidities, and the level of dermatologic monitoring have not been studied.
Regarding CYP2C9 and CYP2C19:
In most individuals maintained at a steady dosage, stable phenytoin serum levels are achieved. There may be wide interindividual variability in phenytoin serum levels with equivalent dosages. Individuals with unusually low levels may be noncompliant or hypermetabolizers of phenytoin.
Unusually high levels result from liver disease, variant CYP2C9 and CYP2C19 alleles, or drug interactions which result in metabolic interference. The individual with large variations in phenytoin serum levels, despite standard doses, presents a difficult clinical problem. Serum level determinations in such individuals may be particularly helpful. As phenytoin is highly protein bound, free phenytoin levels may be altered in individuals whose protein binding characteristics differ from normal.
Please review the complete therapeutic recommendations that are located here: (1).
2018 Summary of recommendations from the Dutch Pharmacogenetics Working Group (DPWG) of the Royal Dutch Association for the Advancement of Pharmacy (KNMP)
HLA-B*1502
The life-threatening cutaneous side effect Stevens-Johnson syndrome/toxic epidermal necrolysis (SJS/TEN) occurs more frequently in patients with this genetic variation. The calculated risk of phenytoin-induced SJS/TEN in patients with HLA-B*15:02 is 0.65%.
- Carefully weigh the risk of SJS/TEN against the benefits
- Avoid phenytoin if an alternative is possible
- Carbamazepine carries a 10-fold higher risk of SJS/TEN for these individuals and is therefore not an alternative.
- A comparable risk has been reported for lamotrigine as for phenytoin. The same applies for oxcarbazepine, but the most severe forms (SJS/TEN overlap and TEN) are not observed with oxcarbazepine.
- If it is not possible to avoid this medication, then advise the individual to report any skin rash immediately (Table 2)
CYP2C9 genotypes *1/*2, *1/*3 and other IMs
Genetic variation reduces conversion of phenytoin to inactive metabolites. This increases the risk of side effects.
Recommendation:
- 1.
The loading dose does not need to be adjusted.
- 2.
For the other doses, use 75% of the standard dose and assess the dose based on effect and serum concentration after 7-10 days.
- 3.
Advise the patient to report if side effects (such as ataxia, nystagmus, slurred speech, sedation or rash) occur.
CYP2C9 genotypes *2/*2 and *2/*3 and other PMs
Genetic variation reduces conversion of phenytoin to inactive metabolites. This increases the risk of side effects.
Recommendation:
- 1.
The loading dose does not need to be adjusted.
- 2.
For the other doses, use 50% of the standard dose and assess the dose based on effect and serum concentration after 7-10 days.
- 3.
Advise the patient to report if side effects (such as ataxia, nystagmus, slurred speech, sedation or rash) occur.
CYP2C9 genotype *3/*3 (PM)
Genetic variation reduces conversion of phenytoin to inactive metabolites. This increases the risk of side effects, including SJS/TEN in Asian patients.
Recommendation:
- 1.
The loading dose does not need to be adjusted.
- 2.
For the other doses, use 40% of the standard dose and assess the dose based on effect and serum concentration after 7-10 days.
- 3.
Advise the patient to report if side effects (such as ataxia, nystagmus, slurred speech,sedation or, especially in Asian individuals, rash) occur.
Please review the complete therapeutic recommendations that are located here (3).
2020 Statement from the Clinical Pharmacogenetics Implementation Consortium (CPIC)
HLA-B*15:02 recommendations
[…] If a individual is phenytoin-naïve and HLA-B*15:02 positive, the individual has an increased risk of SJS/TEN and the recommendation is to consider using an anticonvulsant other than phenytoin unless the benefits of treating the underlying disease clearly outweigh the risks (see Table 3). Carbamazepine and oxcarbazepine should also be avoided if a individual is HLA-B*15:02 positive. Alternative medications such as eslicarbazepine acetate and lamotrigine have limited evidence linking SJS/TEN with the HLA-B*15:02 allele.
[…]
If a individual is phenytoin-naïve and HLA-B*15:02 negative, the individual has a normal risk of phenytoin-induced SJS/TEN and the recommendation is to use phenytoin with dosage adjustments based on CYP2C9 genotype (if known) or standard dosing guidelines (if CYP2C9 genotype is unknown). However, an HLA-B*15:02 negative test does not eliminate the risk of phenytoin-induced SJS/TEN.
CYP2C9 recommendations.
The recommended phenytoin initial or loading and maintenance doses do not need adjustments based on genotype for CYP2C9 NMs and IMs with an AS of ≥1.5. Available evidence does not clearly indicate the extent of dose reduction needed to prevent phenytoin-related toxicities in CYP2C9 IMs with an AS of 1.0 and PMs with an AS of 0 or 0.5. Furthermore, multiple case studies have observed that CYP2C9 PMs are at increased risk for exposure-related phenytoin toxicities, and multiple studies have observed an association between the CYP2C9*3 allele and SJS/TEN. Although presence of the CYP2C9*3 allele is insufficient to predict phenytoin-induced SJS/TEN, these and other data suggest that the risk of SJS/TEN is dose-related and provide an additional rationale for reducing phenytoin dose in CYP2C9 PMs . Thus, our recommendations are conservative given the variability surrounding phenytoin dosing. Based on the doses reported in the pharmacokinetic and pharmacogenetic studies mentioned above and in Table S2, a typical initial or loading dose followed by at least a 25% reduction in the recommended starting maintenance dose may be considered for CYP2C9 IMs with AS of 1.0. Subsequent maintenance doses should be adjusted based on therapeutic drug monitoring and response. For CYP2C9 PMs, use a typical initial or loading dose then consider at least a 50% reduction of starting maintenance dose with subsequent maintenance doses adjusted based on therapeutic drug monitoring and response.
Pediatrics
Much of the evidence (summarized in Table S1) linking HLA-B*15:02 to phenytoin induced SJS/TEN was generated in both children and adults. Therefore, the above recommendation is made regardless of CYP2C9 genotype, individual age, race or ancestry.
Please review the complete therapeutic recommendations that are located here: (2).
Nomenclature of Selected HLA-B Alleles
| Allele name | dbSNP reference identifier for allele location |
|---|---|
| HLA-B*15:02 | Tagged variants cannot be reliably used to detect this allele. Sequencing is the most accurate technique for allele detection. |
For the major histocompatibility complex region, variations in genes such as HLA-B occur across the whole sequence of the gene, not a single locus. Therefore, the HLA-B*15:02 allele is defined by its sequence rather than single coding or protein variations. If there is strong linkage disequilibrium between one or more SNPs and a specific HLA allele, the presence of these SNPs (tag SNPs) may be used for HLA typing in some populations; however, genotyping tagged variants should not be considered diagnostic or equivalent to actual HLA testing. For HLA-B*15:02, rs2844682 and rs3909184 were previously considered the tagged variants (47), however, these variants have shown to be less accurate in other studies (48). Other tagged variants have been suggested, however, the sensitivity and accuracy of these variants to detect the HLA-B*15:02 allele is limited (49, 50). Sequence for the full HLA-B*15:02 allele (and subtypes) can be accessed here.
Guidelines on nomenclature of the HLA system are available from HLA Nomenclature: http://hla
.alleles.org/
Nomenclature of Selected CYP2C9 Alleles
| Common allele name | Alternative names | HGVS reference sequence | dbSNP reference identifier for allele location | |
|---|---|---|---|---|
| Coding | Protein | |||
| CYP2C9*2 | 430C>T Arg144Cys | NM_000771.3:c.430C>T | NP_000762.2:p.Arg144Cys | rs1799853 |
| CYP2C9*3 | 1075A>C Ile359Leu | NM_000771.3:c.1075A>C | NP_000762.2:p.Ile359Leu | rs1057910 |
| CYP2C9*5 | 1080C>G Asp360Glu | NM_000771.3:c.1080C>G | NP_000762.2:p.Asp360Glu | rs28371686 |
| CYP2C9*6 | 817delA Lys273Argfs | NM_000771.3:c.818del | NP_000762.2:p.Lys273Argfs | rs9332131 |
| CYP2C9*8 | 449G>A Arg150His | NM_000771.3:c.449G>A | NP_000762.2:p.Arg150His | rs7900194 |
| CYP2C9*11 | 1003C>T Arg335Trp | NM_000771.3:c.1003C>T | NP_000762.2:p.Arg335Trp | rs28371685 |
Guidelines for the description and nomenclature of gene variations are available from the Human Genome Variation Society (HGVS): http://www
.hgvs.org/content/guidelines Nomenclature for Cytochrome P450 enzymes is available from the Pharmacogene Variation Consortium (PharmVar) https://www
.pharmvar.org/.
Acknowledgments
The authors would like to thank Marga Nijenhuis, PhD, Royal Dutch Pharmacists Association (KNMP), The Hague, The Netherlands; Bernard Esquivel MD, PhD, President of the Latin American Association for Personalized Medicine, Vancouver, BC, Canada; and Jason H. Karnes, PharmD, PhD, BCPS, FAHA, Assistant Professor, Department of Pharmacy Practice & Science, University of Arizona College of Pharmacy; Sarver Heart Center, University of Arizona College of Medicine, Tucson, Arizona, USA for reviewing this summary
First edition:
The author would like to thank Mohamed Nagy, Clinical Pharmacist, Head of the Personalised Medication Management Unit, Department of Pharmaceutical Services, Children's Cancer Hospital, Cairo, Egypt; Emily K. Pauli, Director of Research, Clearview Cancer Institute, Huntsville, AL, USA; Michael A. Rogawski, Professor of Neurology, University of California, Davis, CA, USA; and Stuart A. Scott, Assistant Professor of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, USA for reviewing this summary.
Version History
To view the previous version of this chapter, published on 22 September 2016, please click here.
References
- 1.
- PHENYTOIN suspension [package insert]. Morton Grove, IL: Morton Grove Pharmaceuticals Inc; 2019. Available from: https://dailymed
.nlm .nih.gov/dailymed/drugInfo .cfm?setid=efd93f07-818b-41ae-abd6-49ec5175311a. - 2.
- Karnes J.H., Rettie A.E., Somogyi A.A., Huddart R., et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C9 and HLA-B Genotypes and Phenytoin Dosing: 2020 Update. Clin Pharmacol Ther. 2021;109(2):302–309. [PMC free article: PMC7831382] [PubMed: 32779747]
- 3.
- Royal Dutch Pharmacists Association (KNMP). Dutch Pharmacogenetics Working Group (DPWG). Pharmacogenetic Guidelines [Internet]. Netherlands. Phenytoin – HLA-B*1502 [Cited July 2020]. Available from: http://kennisbank
.knmp.nl. - 4.
- Caudle K.E., Dunnenberger H.M., Freimuth R.R., Peterson J.F., et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med. 2017;19(2):215–223. [PMC free article: PMC5253119] [PubMed: 27441996]
- 5.
- Royal Dutch Pharmacists Association (KNMP). Dutch Pharmacogenetics Working Group (DPWG). Pharmacogenetic Guidelines [Internet]. Netherlands. Phenytoin – CYP2C9 [Cited July 2020]. Available from: http://kennisbank
.knmp.nl. - 6.
- Catterall W.A. Molecular properties of brain sodium channels: an important target for anticonvulsant drugs. Adv Neurol. 1999;79:441–56. [PubMed: 10514834]
- 7.
- Lipkind G.M., Fozzard H.A. Molecular model of anticonvulsant drug binding to the voltage-gated sodium channel inner pore. Mol Pharmacol. 2010;78(4):631–8. [PMC free article: PMC2981395] [PubMed: 20643904]
- 8.
- Segal M.M., Douglas A.F. Late sodium channel openings underlying epileptiform activity are preferentially diminished by the anticonvulsant phenytoin. J Neurophysiol. 1997;77(6):3021–34. [PubMed: 9212254]
- 9.
- Birth Defects: Data & Statistics. Centers for Disease Control and Prevention (CDC); Available from: http://www
.cdc.gov/ncbddd /birthdefects/data.html. - 10.
- Hill D.S., Wlodarczyk B.J., Palacios A.M., Finnell R.H. Teratogenic effects of antiepileptic drugs. Expert Rev Neurother. 2010;10(6):943–59. [PMC free article: PMC2970517] [PubMed: 20518610]
- 11.
- Mullan K.A., Anderson A., Illing P.T., Kwan P., et al. HLA-associated antiepileptic drug-induced cutaneous adverse reactions. HLA. 2019;93(6):417–435. [PubMed: 30895730]
- 12.
- Svensson C.K., Cowen E.W., Gaspari A.A. Cutaneous drug reactions. Pharmacol Rev. 2001;53(3):357–79. [PubMed: 11546834]
- 13.
- Nomenclature for Factors of the HLA System: HLA Alleles [Cited 23 June 2016]. Available from: http://hla
.alleles.org/alleles/index .html. - 14.
- Choo S.Y. The HLA system: genetics, immunology, clinical testing, and clinical implications. Yonsei Med J. 2007;48(1):11–23. [PMC free article: PMC2628004] [PubMed: 17326240]
- 15.
- Chung W.H., Hung S.I., Hong H.S., Hsih M.S., et al. Medical genetics: a marker for Stevens-Johnson syndrome. Nature. 2004;428(6982):486. [PubMed: 15057820]
- 16.
- Amstutz U., Shear N.H., Rieder M.J., Hwang S., et al. Recommendations for HLA-B*15:02 and HLA-A*31:01 genetic testing to reduce the risk of carbamazepine-induced hypersensitivity reactions. Epilepsia. 2014;55(4):496–506. [PubMed: 24597466]
- 17.
- Locharernkul C., Loplumlert J., Limotai C., Korkij W., et al. Carbamazepine and phenytoin induced Stevens-Johnson syndrome is associated with HLA-B*1502 allele in Thai population. Epilepsia. 2008;49(12):2087–91. [PubMed: 18637831]
- 18.
- Hung S.I., Chung W.H., Liu Z.S., Chen C.H., et al. Common risk allele in aromatic antiepileptic-drug induced Stevens-Johnson syndrome and toxic epidermal necrolysis in Han Chinese. Pharmacogenomics. 2010;11(3):349–56. [PubMed: 20235791]
- 19.
- TEGRETOL (carbamazepine) tablet [package insert]. East Hanover, New Jersey 07936: Corporation, N.P.; 2011. Available from: http://dailymed
.nlm.nih .gov/dailymed/lookup .cfm?setid=8d409411-aa9f-4f3a-a52c-fbcb0c3ec053. - 20.
- Leckband S.G., Kelsoe J.R., Dunnenberger H.M., George A.L. Jr, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for HLA-B genotype and carbamazepine dosing. Clin Pharmacol Ther. 2013;94(3):324–8. [PMC free article: PMC3748365] [PubMed: 23695185]
- 21.
- Chung W.H., Hung S.I., Chen Y.T. Genetic predisposition of life-threatening antiepileptic-induced skin reactions. Expert Opin Drug Saf. 2010;9(1):15–21. [PubMed: 20001755]
- 22.
- Puangpetch A., Koomdee N., Chamnanphol M., Jantararoungtong T., et al. HLA-B allele and haplotype diversity among Thai patients identified by PCR-SSOP: evidence for high risk of drug-induced hypersensitivity. Front Genet. 2014;5:478. [PMC free article: PMC4302987] [PubMed: 25657656]
- 23.
- Nguyen D.V., Chu H.C., Nguyen D.V., Phan M.H., et al. HLA-B*1502 and carbamazepine-induced severe cutaneous adverse drug reactions in Vietnamese. Asia Pac Allergy. 2015;5(2):68–77. [PMC free article: PMC4415182] [PubMed: 25938071]
- 24.
- Chong K.W., Chan D.W., Cheung Y.B., Ching L.K., et al. Association of carbamazepine-induced severe cutaneous drug reactions and HLA-B*1502 allele status, and dose and treatment duration in paediatric neurology patients in Singapore. Arch Dis Child. 2014;99(6):581–4. [PubMed: 24225276]
- 25.
- Hung S.I., Chung W.H., Jee S.H., Chen W.C., et al. Genetic susceptibility to carbamazepine-induced cutaneous adverse drug reactions. Pharmacogenet Genomics. 2006;16(4):297–306. [PubMed: 16538176]
- 26.
- Lonjou C., Thomas L., Borot N., Ledger N., et al. A marker for Stevens-Johnson syndrome ...: ethnicity matters. Pharmacogenomics J. 2006;6(4):265–8. [PubMed: 16415921]
- 27.
- Alfirevic A., Jorgensen A.L., Williamson P.R., Chadwick D.W., et al. HLA-B locus in Caucasian patients with carbamazepine hypersensitivity. Pharmacogenomics. 2006;7(6):813–8. [PubMed: 16981842]
- 28.
- Kaniwa N., Saito Y., Aihara M., Matsunaga K., et al. HLA-B locus in Japanese patients with anti-epileptics and allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis. Pharmacogenomics. 2008;9(11):1617–22. [PubMed: 19018717]
- 29.
- Mehta T.Y., Prajapati L.M., Mittal B., Joshi C.G., et al. Association of HLA-B*1502 allele and carbamazepine-induced Stevens-Johnson syndrome among Indians. Indian J Dermatol Venereol Leprol. 2009;75(6):579–82. [PubMed: 19915237]
- 30.
- Wu X.T., Hu F.Y., An D.M., Yan B., et al. Association between carbamazepine-induced cutaneous adverse drug reactions and the HLA-B*1502 allele among patients in central China. Epilepsy Behav. 2010;19(3):405–8. [PubMed: 20833111]
- 31.
- Van Booven D., Marsh S., McLeod H., Carrillo M.W., et al. Cytochrome P450 2C9-CYP2C9. Pharmacogenet Genomics. 2010;20(4):277–81. [PMC free article: PMC3201766] [PubMed: 20150829]
- 32.
- Gupta A., Zheng L., Ramanujam V., Gallagher J. Novel Use of Pharmacogenetic Testing in the Identification of CYP2C9 Polymorphisms Related to NSAID-Induced Gastropathy. Pain Med. 2015;16(5):866–9. [PubMed: 25585969]
- 33.
- Sistonen J., Fuselli S., Palo J.U., Chauhan N., et al. Pharmacogenetic variation at CYP2C9, CYP2C19, and CYP2D6 at global and microgeographic scales. Pharmacogenet Genomics. 2009;19(2):170–9. [PubMed: 19151603]
- 34.
- Solus J.F., Arietta B.J., Harris J.R., Sexton D.P., et al. Genetic variation in eleven phase I drug metabolism genes in an ethnically diverse population. Pharmacogenomics. 2004;5(7):895–931. [PubMed: 15469410]
- 35.
- Lee C.R., Goldstein J.A., Pieper J.A. Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002;12(3):251–63. [PubMed: 11927841]
- 36.
- Liao K., Liu Y., Ai C.Z., Yu X., et al. The association between CYP2C9/2C19 polymorphisms and phenytoin maintenance doses in Asian epileptic patients: A systematic review and meta-analysis. Int J Clin Pharmacol Ther. 2018;56(7):337–346. [PubMed: 29628024]
- 37.
- Fohner A.E., Ranatunga D.K., Thai K.K., Lawson B.L., et al. Assessing the clinical impact of CYP2C9 pharmacogenetic variation on phenytoin prescribing practice and patient response in an integrated health system. Pharmacogenet Genomics. 2019;29(8):192–199. [PMC free article: PMC6989102] [PubMed: 31461080]
- 38.
- Calderon-Ospina C.A., Galvez J.M., Lopez-Cabra C., Morales N., et al. Possible Genetic Determinants of Response to Phenytoin in a Group of Colombian Patients With Epilepsy. Front Pharmacol. 2020;11:555. [PMC free article: PMC7221122] [PubMed: 32457604]
- 39.
- Wen Y.F., Culhane-Pera K.A., Thyagarajan B., Bishop J.R., et al. Potential Clinical Relevance of Differences in Allele Frequencies Found within Very Important Pharmacogenes between Hmong and East Asian Populations. Pharmacotherapy. 2020;40(2):142–152. [PubMed: 31884695]
- 40.
- Fohner A.E., Rettie A.E., Thai K.K., Ranatunga D.K., et al. Associations of CYP2C9 and CYP2C19 Pharmacogenetic Variation with Phenytoin-Induced Cutaneous Adverse Drug Reactions. Clin Transl Sci. 2020;13(5):1004–1009. [PMC free article: PMC7485959] [PubMed: 32216088]
- 41.
- Chang W.C., Hung S.I., Carleton B.C., Chung W.H. An update on CYP2C9 polymorphisms and phenytoin metabolism: implications for adverse effects. Expert Opin Drug Metab Toxicol. 2020;16(8):723–734. [PubMed: 32510242]
- 42.
- Yampayon K., Sukasem C., Limwongse C., Chinvarun Y., et al. Influence of genetic and non-genetic factors on phenytoin-induced severe cutaneous adverse drug reactions. Eur J Clin Pharmacol. 2017;73(7):855–865. [PubMed: 28391407]
- 43.
- Su S.C., Chen C.B., Chang W.C., Wang C.W., et al. HLA Alleles and CYP2C9*3 as Predictors of Phenytoin Hypersensitivity in East Asians. Clin Pharmacol Ther. 2019;105(2):476–485. [PubMed: 30270535]
- 44.
- Hikino K., Ozeki T., Koido M., Terao C., et al. HLA-B*51:01 and CYP2C9*3 Are Risk Factors for Phenytoin-Induced Eruption in the Japanese Population: Analysis of Data From the Biobank Japan Project. Clin Pharmacol Ther. 2020;107(5):1170–1178. [PubMed: 31646624]
- 45.
- Sukasem C., Sririttha S., Tempark T., Klaewsongkram J., et al. Genetic and clinical risk factors associated with phenytoin-induced cutaneous adverse drug reactions in Thai population. Pharmacoepidemiol Drug Saf. 2020;29(5):565–574. [PubMed: 32134161]
- 46.
- Pratt V.M., Cavallari L.H., Del Tredici A.L., Hachad H., et al. Recommendations for Clinical CYP2C9 Genotyping Allele Selection: A Joint Recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2019;21(5):746–755. [PMC free article: PMC7057225] [PubMed: 31075510]
- 47.
- de Bakker P.I., McVean G., Sabeti P.C., Miretti M.M., et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38(10):1166–72. [PMC free article: PMC2670196] [PubMed: 16998491]
- 48.
- Zhu G.D., Brenton A.A., Malhotra A., Riley B.J., et al. Genotypes at rs2844682 and rs3909184 have no clinical value in identifying HLA-B*15:02 carriers. Eur J Clin Pharmacol. 2015;71(8):1021–3. [PubMed: 26036218]
- 49.
- He Y., Hoskins J.M., Clark S., Campbell N.H., et al. Accuracy of SNPs to predict risk of HLA alleles associated with drug-induced hypersensitivity events across racial groups. Pharmacogenomics. 2015;16(8):817–24. [PubMed: 26083016]
- 50.
- Fang H., Xu X., Kaur K., Dedek M., et al. A Screening Test for HLA-B (*)15:02 in a Large United States Patient Cohort Identifies Broader Risk of Carbamazepine-Induced Adverse Events. Front Pharmacol. 2019;10:149. [PMC free article: PMC6443844] [PubMed: 30971914]
Footnotes
- 1
The FDA labels specific drug formulations. We have substituted the generic names for any drug labels in this excerpt. The FDA may not have labeled all formulations containing the generic drug.
- Introduction
- Drug: Phenytoin
- The HLA Gene Family
- Gene: HLA-B*15:02
- Gene: CYP2C9
- Linking HLA-B and CYP2C9 Genetic Variation with the Risk of Side Effects and Treatment Response
- The HLA-B and CYP2C9 Gene Interactions with Medications Used for Additional Indications
- Genetic Testing
- Therapeutic Recommendations based on Genotype
- Nomenclature of Selected HLA-B Alleles
- Nomenclature of Selected CYP2C9 Alleles
- Acknowledgments
- Version History
- References
- Abacavir Therapy and HLA-B*57:01 Genotype
- Allopurinol Therapy and HLA-B*58:01 Genotype
- Carbamazepine Therapy and HLA Genotype
- Celecoxib Therapy and CYP2C9 Genotype
- Dronabinol Therapy and CYP2C9 Genotype
- Flurbiprofen Therapy and CYP2C9 Genotype
- Lesinurad Therapy and CYP2C9 Genotype
- Piroxicam Therapy and CYP2C9 Genotype
- Prasugrel Therapy and CYP Genotype
- Siponimod Therapy and CYP2C9 Genotype
- Warfarin Therapy and VKORC1 and CYP Genotype
- Review Carbamazepine Therapy and HLA Genotype.[Medical Genetics Summaries. 2012]Review Carbamazepine Therapy and HLA Genotype.Dean L. Medical Genetics Summaries. 2012
- Review Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C9 and HLA-B Genotypes and Phenytoin Dosing: 2020 Update.[Clin Pharmacol Ther. 2021]Review Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C9 and HLA-B Genotypes and Phenytoin Dosing: 2020 Update.Karnes JH, Rettie AE, Somogyi AA, Huddart R, Fohner AE, Formea CM, Ta Michael Lee M, Llerena A, Whirl-Carrillo M, Klein TE, et al. Clin Pharmacol Ther. 2021 Feb; 109(2):302-309. Epub 2020 Sep 6.
- Review Warfarin Therapy and VKORC1 and CYP Genotype.[Medical Genetics Summaries. 2012]Review Warfarin Therapy and VKORC1 and CYP Genotype.Dean L. Medical Genetics Summaries. 2012
- Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and HLA-B genotypes and phenytoin dosing.[Clin Pharmacol Ther. 2014]Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and HLA-B genotypes and phenytoin dosing.Caudle KE, Rettie AE, Whirl-Carrillo M, Smith LH, Mintzer S, Lee MT, Klein TE, Callaghan JT, Clinical Pharmacogenetics Implementation Consortium. Clin Pharmacol Ther. 2014 Nov; 96(5):542-8. Epub 2014 Aug 6.
- Review An update on CYP2C9 polymorphisms and phenytoin metabolism: implications for adverse effects.[Expert Opin Drug Metab Toxicol...]Review An update on CYP2C9 polymorphisms and phenytoin metabolism: implications for adverse effects.Chang WC, Hung SI, Carleton BC, Chung WH. Expert Opin Drug Metab Toxicol. 2020 Aug; 16(8):723-734. Epub 2020 Jul 16.
- Phenytoin Therapy and HLA-B*15:02 and CYP2C9 Genotype - Medical Genetics Summari...Phenytoin Therapy and HLA-B*15:02 and CYP2C9 Genotype - Medical Genetics Summaries
Your browsing activity is empty.
Activity recording is turned off.
See more...
