All Medical Genetics Summaries content, except where otherwise noted, is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license which permits copying, distribution, and adaptation of the work, provided the original work is properly cited and any changes from the original work are properly indicated. Any altered, transformed, or adapted form of the work may only be distributed under the same or similar license to this one.
NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Pratt VM, Scott SA, Pirmohamed M, et al., editors. Medical Genetics Summaries [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2012-.
Introduction
Warfarin (brand name Coumadin) is an anticoagulant (blood thinner). Warfarin acts by inhibiting the synthesis of vitamin K-dependent clotting factors and is used in the prevention and treatment of various thrombotic disorders. Warfarin is a drug with narrow therapeutic index; thus, a small change in its plasma levels may result in concentration dependent adverse drug reactions or therapeutic failure. Therefore, the dose of warfarin must be tailored for each patient according to the patient’s response, measured as INR (International Normalized Ratio), and the condition being treated.
There is a wide inter-individual variability in the dose of warfarin required to achieve target anticoagulation, and the time it takes to reach target INR. Approximately half of this variability is known to be caused by clinical or lifestyle factors (e.g., a patient’s age, weight, BMI, gender, smoking status, existing conditions, and concomitant medications) and by genetic factors (known genetic factors include variants in the VKORC1, CYP2C9, CYP4F2 genes, and the rs12777823 variant in the CYP2C gene cluster on chromosome 10) (1).
The VKORC1 and CYP2C9 genotypes are the most important known genetic determinants of warfarin dosing. Warfarin targets VKORC1, an enzyme involved in vitamin K recycling. A common variant, VKORC1, c.-1639G>A, is associated with an increased sensitivity to warfarin and lower dose requirements. The CYP2C9 enzyme metabolizes warfarin and the variants CYP2C9*2 and *3, are also associated with lower dose requirements.
The FDA-approved drug label for warfarin states that CYP2C9 and VKORC1 genotype information, when available, can assist in the selection of the initial dose of warfarin. The label provides 2 sets of warfarin dosing recommendations, for when the CYP2C9 and VKORC1 genotypes are either known (Table 1) or not known (taking into account clinical factors, the initial dose of warfarin is usually 2–5 mg once daily) (1).
In addition, the Dutch Pharmacogenetics Working Group (DPWG) of the Royal Dutch Association for the Advancement of Pharmacy (KNMP) has published recommendations for the initial standard dose of warfarin. A dose reduction is recommended for individuals who are CYP2C9 poor and intermediate metabolizers (with the exception of intermediate metabolizers with the CYP2C9*1/*2 genotype, no dose change is required), and a dose reduction is recommended for individuals who carry 2 copies of the variant VKORC1 A allele (VKORC1, c.-1639G>A/A) (Table 2) (2, 3).
Recently, genetic variation in the CYP4F2 gene, and a variant near the CYP2C gene cluster, rs12777823, have been associated with influencing warfarin therapy. The CYP4F2*3 variant is associated with a modest increase in warfarin dose requirements in individuals with European or Asian ancestry, while in individuals with African ancestry, the rs12777823 A/G or A/A genotype is associated with decreased warfarin dose requirements.
The 2017 Update of the Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing, provides warfarin dosing recommendations for adults with and without African ancestry, and also for pediatric patients (see Therapeutic Recommendations). CPIC recommends that these dosing guidelines are applied after a warfarin dose has been calculated using a validated pharmacogenetic algorithm, which includes genotype information for VKORC1, c.-1639G>A and CYP2C9*2 and *3 (Figure 1) (4)
Table 1.
The FDA (2017) Drug Label for Warfarin. Three Ranges of Expected Maintenance Warfarin Doses based on CYP2C9 and VKORC1 Genotype.
| VKORC1 | CYP2C9 | |||||
|---|---|---|---|---|---|---|
| *1/*1 | *1/*2 | *1/*3 | *2/*2 | *2/*3 | *3/*3 | |
| GG | 5–7 mg | 5–7 mg | 3–4 mg | 3–4 mg | 3–4 mg | 0.5–2 mg |
| AG | 5–7mg | 3–4 mg | 3–4 mg | 3–4 mg | 0.5–2 mg | 0.5–2 mg |
| AA | 3–4 mg | 3–4 mg | 0.5–2 mg | 0.5–2 mg | 0.5–2 mg | 0.5–2 mg |
Ranges are derived from multiple published clinical studies. The VKORC1, c.–1639G>A (rs9923231) variant is used in this table. Other co-inherited VKORC1 variants may also be important determinants of warfarin dose. Patients with CYP2C9 *1/*3, *2/*2, *2/*3, and *3/*3 may require more prolonged time (>2–4 weeks) to achieve a maximum international normalized ratio (INR) effect for a given dosage regimen than patients without these CYP variants.
Please see Therapeutic Recommendations based on Genotype for more information. This table is adapted from the FDA-approved drug label for warfarin (1).
Table 2.
The DPWG (2017) Recommendations for Warfarin and CYP2C9 and VKORC1 Genotype.
| Phenotype/diplotype | Recommendation |
|---|---|
| CYP2C9 IM | Use 65% of the standard initial dose |
| CYP2C9 PM | Use 20% of the standard initial dose |
| CYP2C9*1/*2 | No action is required for this gene-drug interaction. |
| CYP2C9*1/*3 | Use 65% of the standard initial dose |
| CYP2C9*2/*2 | Use 65% of the standard initial dose |
| CYP2C9*2/*3 | Use 45% of the standard initial dose |
| CYP2C9*3/*3 | Use 20% of the standard initial dose |
| VKORC1 C/T | No action is required for this gene-drug interaction |
| VKORC1 T/T | Use 60% of the standard initial dose |
Note: VKORC1 1173C>T is equivalent to c.-1639G>A. Therefore:
“VKORC1 CT” corresponds to VKORC1, c.-1639 G/A
“VKORC1 TT” corresponds to VKORC1, c.-1639 A/APlease see Therapeutic Recommendations based on Genotype for more information from the Dutch Pharmacogenetics Working Group (DPWG). Table is adapted from (2, 3).
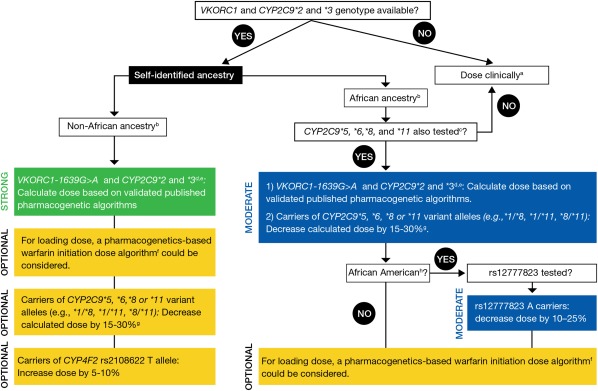
Figure 1.
The CPIC (2017) Dosing Recommendations for Warfarin Dosing based on Genotype for Adult Patients. (a) “Dose clinically” means to dose without genetic information, which may include use of a clinical dosing algorithm or standard dose approach. (b) Data strongest for European and East Asian ancestry populations and consistent in other populations. (c) 45–50% of individuals with self‐reported African ancestry carry CYP2C9*5, *6, *8, *11, or rs12777823. If CYP2C9*5, *6, *8, and *11 were not tested, dose warfarin clinically. Note: these data derive primarily from African-Americans, who are largely from West Africa. It is unknown if the same associations are present for those from other parts of Africa. (d) Most algorithms are developed for the target INR 2‐3. (e) Consider an alternative agent in individuals with genotypes associated with CYP2C9 poor metabolism (e.g., CYP2C9*3/*3, *2/*3, *3/*3) or both increased sensitivity (VKORC1 A/G or A/A) and CYP2C9 poor metabolism. (f) See the EU‐PACT trial for pharmacogenetics‐based warfarin initiation (loading) dose algorithm with the caveat that the loading dose algorithm has not been specifically tested or validated in populations of African ancestry. (g) Larger dose reduction might be needed in variant homozygotes (i.e., 20–40%). (h) African-American refers to individuals mainly originating from West Africa.
This figure is adapted from (4). Please see Therapeutic Recommendations based on Genotype for more information from CPIC.
Drug: Warfarin
Warfarin is an anticoagulant used in the prevention and treatment of venous thrombosis, pulmonary embolism, and the complications associated with atrial fibrillation and/or cardiac valve replacement. Warfarin is sometimes prescribed to reduce the risk of stroke after a myocardial infarction (MI).
Warfarin has no direct effect on an established thrombus. However, once a thrombus has occurred (e.g., deep venous thrombosis), the goal of warfarin therapy is to prevent further extension of the formed clot and to prevent secondary thromboembolic complications that may be fatal (e.g., pulmonary embolism).
Warfarin is a teratogen – an agent that can cause abnormalities in a developing fetus. Therefore, warfarin use in pregnancy is contraindicated, except in women with mechanical heart valves who have a particularly high risk of thromboembolism. If warfarin is used in pregnancy, or if a patient becomes pregnant while taking warfarin, she should be informed of the potential risks to the fetus (1).
Warfarin exposure in pregnancy can cause fetal death, neonatal death, and warfarin syndrome - a pattern of developmental abnormalities that most commonly affect bone and cartilage, causing nasal hypoplasia, and a “stippled” appearance to the ends of long bones. The risk of warfarin teratogenicity appears to be greatest between the 6th and 12th week of pregnancy, but toxicity before or after this period is still possible (5, 6).
Warfarin exerts its anticoagulant effect by inhibiting the enzyme encoded by VKORC1, which catalyzes the conversion of vitamin K epoxide to the active reduced form of vitamin K, vitamin K hydroquinone. Vitamin K hydroquinone is an essential cofactor in the synthesis of several clotting factors and decreased availability of vitamin K hydroquinone leads to decreased activity of the clotting factors II, VII, IX, and X, and the anticoagulant proteins C and S (7).
Warfarin is administered as a racemic mixture of the R- and S- stereoisomers. (S)-warfarin is 2–5 times more potent than (R)-warfarin and is mainly metabolized by CYP2C9. (R)-warfarin is mainly metabolized by other cytochrome P450 enzymes (8).
The initial and maintenance doses of warfarin must be tailored to each patient, and monitoring of the international normalized ratio (INR) should be performed in all patients treated with warfarin. The INR is a standardized measurement of prothrombin time, which is the time it takes for blood to clot. In healthy individuals, the INR is approximately one (range: 0.8–1.1). The goal of warfarin therapy is to achieve an INR in a target range for the condition being treated (most commonly 2–3).
The FDA-approved drug label for warfarin carries a boxed warning cautioning of the risk of bleeding, which can be fatal. Bleeding is more likely to occur within the first month, and risk factors include a high intensity of anticoagulation (INR greater than 4), age greater than or equal to 65, and a history of highly variable INRs. Other serious adverse events associated with warfarin therapy include necrosis of the skin and other tissues, particularly when used prematurely to manage thrombosis associated with heparin-induced thrombocytopenia (HIT).
Since warfarin is a drug with a narrow therapeutic index, an optimal starting dose may reduce the time taken to reach a stable INR and reduce the risk of having either a high INR (with a risk of bleeding) or a low INR (with a risk of thrombosis). Known factors that influence an individual’s response to the initial dose of warfarin include clinical and lifestyle factors (e.g., age, race, body weight, height, gender, concomitant medications—including those that compete for binding to albumin, comorbidities, diet, nutritional status) and genetic factors (e.g., CYP2C9 and VKORC1 genotypes). Therefore, the initial dose should be modified to take into account these and any additional patient-specific factors that may influence warfarin dose requirement.
The FDA-approved drug label for warfarin suggests considering a lower initial and maintenance dose of warfarin for elderly and/or debilitated patients, and in Asian patients. The drug label recommends against the routine use of loading doses because this practice may increase hemorrhagic and other complications and does not offer more rapid protection against clot formation. However, loading doses are used in practice, and are addressed in CPIC recommendations (4).
The drug label also provides a dosing table of expected maintenance daily doses of warfarin based on CYP2C9 and VKORC1 genotypes (Table 1). The label states that if the patient’s CYP2C9 and/or VKORC1 genotypes are known, to consider these doses when selecting the initial dose of warfarin. However, CPIC states that genetics-based algorithms, such as the International Warfarin Pharmacogenetics Consortium (IWPC), predicts warfarin dose better than the table in the drug label (9).
CPIC has provided dosing recommendations that take into account whether the patients VKORC1 and CYP2C9*2 and *3 genotype is available, and a patient's self-identified ancestry (African ancestry or non-African ancestry). For patients with African ancestry, the presence of CYP2C9*5, *6, *8, and *11 alleles, and rs12777823 are also taken into account (4).
Gene: VKORC1
Genetic variation in the VKORC1 gene is the most important known genetic factor that influences warfarin dosing. Pharmacogenomic algorithms for warfarin dosing routinely include testing for VKORC1.
The VKORC1 gene encodes the vitamin K epoxide reductase enzyme, which catalyzes the rate-limiting step in vitamin K recycling (converting vitamin K epoxide to vitamin K). This enzyme is also the drug target for warfarin.
A common non-coding variant, VKORC1, c.-1639G>A (rs9923231), is associated with an increased sensitivity to warfarin and lower dose requirements (10). The polymorphism occurs in the promoter region of VKORC1 and is thought to alter a transcription factor binding site, leading to lower protein expression. As a result, patients starting warfarin therapy who are carrying at least one “A “allele at -1639 locus require lower initial and maintenance doses compared with patients carrying a G/G genotype at this locus.
The VKORC1, c.−1639G>A allele frequency varies among different ethnic groups. It is the major allele (around 90%) in Asian populations and may be one of the contributing factors for lower warfarin dosing requirements often observed in patients of Asian descent. It is also common in Caucasians (around 40%) and African-Americans (around 14%) (11-13).
Less commonly, missense mutations in VKORC1 can lead to warfarin resistance and higher dose requirements (14, 15).
The Cytochrome P450 Superfamily
The cytochrome P450 superfamily (CYP450) is a large and diverse group of enzymes that form the major system for metabolizing or detoxifying lipids, hormones, toxins, and drugs in the liver. The CYP450 genes are very polymorphic and can result in reduced, absent, or increased enzyme activity.
CYP450 isozymes involved in the metabolism of warfarin include CYP2C9, CYP3A4, and CYP1A2. The more potent warfarin S-enantiomer is metabolized by CYP2C9 while the R-enantiomer is metabolized by CYP1A2 and CYP3A4. The FDA-approved drug label for warfarin states that drugs that inhibit or induce CYP2C9, CYP1A2, and/or CYP3A4 can influence warfarin exposure and increase or decrease the INR.
The influence of genetic variants in CYP2C9, CYP4F2, and the CYP2C gene cluster, is discussed below.
Gene: CYP2C9
Genetic variation in the CYP2C9 gene is a well-known genetic factor that influences warfarin dosing. Pharmacogenomic algorithms for warfarin dosing routinely include testing for CYP2C9.
The CYP2C9 gene is highly polymorphic, with over 60 star (*) alleles described and currently cataloged at the Pharmacogene Variation (PharmVar) Consortium. The CYP2C9*1 allele is the wild-type allele, and is associated with normal enzyme activity and the normal metabolizer phenotype.
The frequencies of the CYP2C9 alleles vary between different ethnic groups (16-18). In individuals of European descent, the 2 most common variant alleles associated with reduced enzyme activity are CYP2C9*2 (c.430C>T; rs1799853) and *3 (c.1075A>C; rs1057910). The *2 allele is more common in Caucasian (10-20%) than African (0-6%) populations (19). The *3 allele is less common (<10% in most populations), but rare in African populations (20).
Compared to normal metabolizers, individuals of European ancestry who carry one or two copies of *2 or *3 are more sensitive to warfarin—they require lower doses and are at a greater risk of bleeding during warfarin initiation (21-25).
In African-Americans, CYP2C9*5, *6, *8, and *11 variant alleles contribute to the variability in patient response to warfarin (26). These alleles are found more commonly in individuals with African ancestry, and collectively, are more common than the CYP2C9*2 and *3 alleles.
Gene: CYP4F2
The CYP4F2 enzyme is involved in the metabolism of vitamin K in the liver. It is a vitamin K oxidase enzyme and is an important counterpart to VKORC1, a vitamin K reductase enzyme. While VKORC1 catalyzes vitamin K recycling, CYP4F2 limits the excessive accumulation of vitamin K in the liver by catalyzing the production of hydroxylated vitamin K, which is removed from the vitamin K cycle (27).
A genetic variant CYP4F2*3 (c.1297C>T, rs2108622), has been found to influence warfarin dosing. The frequency of the variant T allele is approximately 30% in Caucasians and Asians, and approximately 7% in African-Americans (28).
The CYP4F2 enzyme with an amino acid change due to missense *3 allele is thought to be less active, leading to a rise in hepatic vitamin K. This leads to a higher dose of warfarin being required to achieve therapeutic anticoagulation (by inhibiting vitamin K-dependent clotting factors) (27).
The first studies of CYP4F2 and warfarin dosing reported that Caucasian individuals with the variant rs2108622 TT genotype required approximately 1 mg/day more warfarin than individuals with the rs2108622 CC genotype (28). Two more recent meta-analyses concluded that “T carriers” (individuals with CT or TT genotypes) require approximately an 8–11% increase in warfarin dose, compared to CC individuals. However, data did not support CYP4F2 influencing warfarin requirements in African-Americans (29, 30).
The inclusion of this CYP4F2 variant in warfarin dosing models moderately improves the accuracy of warfarin dose prediction for individuals of European or Asian ancestry, but not for individuals of African ancestry. Accordingly, CPIC recommends that the dose of warfarin should be increased by 5–10% in non-African-American individuals who carry the CYP4F2*3 variant (optional recommendation). CPIC makes no recommendation for African-Americans, stating that data do not support an impact of this variant on warfarin dosing in those of African ancestry (moderate recommendation) (4, 29, 30).
Gene: CYP2C rs12777823
The genetic variant rs12777823, located in the CYP2C gene cluster, is a non-coding variant associated with reduced warfarin dose requirements in African-Americans. The rs12777823 variant was associated with altered warfarin clearance, and individuals with this variant require a lower maintenance dose of warfarin than individuals who do not have this variant (31).
The rs12777823 variant is common in African-Americans (allele frequency 25%) and is also common in other populations; for example, Japanese (32%), and European (15%). However, the association with warfarin dose requirement has only been found for African-Americans: individuals who are heterozygous for the rs12777823 A allele require a dose reduction of warfarin by 7 mg/week, and individuals who are homozygous for the rs12777823 A allele require a dose reduction of warfarin by 9 mg/week (31). Data are lacking for the role of rs12777823 and warfarin response in other populations.
Current pharmacogenomic dosing algorithms do not include rs12777823 status, but analysis has shown that the addition of this variant improves the dosing algorithm published by the IWPC by 21% for African-Americans (31).
CPIC has stated that for African-Americans, a dose reduction of 10–25% in individuals with the rs12777823 A/G or A/A genotype is recommended (moderate recommendation). For non-African-Americans, CPIC recommends that rs12777823 should not be considered, even if the result is available (4).
Genetic Testing
The NIH’s Genetic Testing Registry (GTR) provides a list of tests for “warfarin response,” and the VKORC1, CYP2C9, and CYP4F2 genes.
The VKORC1 and CYP2C9 genotypes are important genetic determinants of warfarin dosing. The contribution of VKORC1 to the variation in dose requirement is larger (approximately 30%) than the contribution of CYP2C9 (usually less than 10%) (32). The variants that are routinely tested for are CYP2C9*2, CYP2C9*3, and VKORC1, c.−1639G>A. These variants are used in the FDA table to guide therapy, and also in the IWPC algorithm.
Currently, routine lab tests do not test for the presence of rs12777823. Other variants that are not routinely tested for include the CYP2C9*5, *6, *8 and *11 alleles, the genes CYP4F2, EPHX1, and GGCX (which all have a role in the vitamin K cycle), and the gene CALU (a cofactor in the VKOR complex) (26, 33).
In African-Americans, the influence of the CYP2C9*5, *6, *8 and *11 alleles are thought to be as significant as the influence of the CYP2C9*2 and*3 alleles on warfarin dosing in Caucasians. Requesting testing of these additional CYP2C9 alleles, and including these genotypes in an expanded dosing algorithm improves warfarin dose prediction in African-Americans, while maintaining high performance in European-Americans (34).
Individuals who are most likely to benefit from genetic testing are those who have yet to start warfarin therapy. However, genotype-guided warfarin dosing is controversial and is generally not carried out preemptively. Some studies have reported that, in general, the current use of genotype-guided dosing algorithms did not improve anticoagulation control in the first few weeks of warfarin therapy (35-41); however, a recent study found genotype-guided warfarin dosing did improve the safety of starting warfarin, compared to clinically guided dosing (42).
Therapeutic Recommendations based on Genotype
This section contains excerpted1 information on gene-based dosing recommendations. Neither this section nor other parts of this review contain the complete recommendations from the sources.
2017 Statement from the US Food and Drug Administration (FDA)
Initial and Maintenance Dosing
The appropriate initial dosing of warfarin sodium tablets varies widely for different patients. Not all factors responsible for warfarin dose variability are known, and the initial dose is influenced by:
- Clinical factors including age, race, body weight, sex, concomitant medications, and comorbidities
- Genetic factors (CYP2C9 and VKORC1 genotypes)
Select the initial dose based on the expected maintenance dose, taking into account the above factors. Modify this dose based on consideration of patient-specific clinical factors. Consider lower initial and maintenance doses for elderly and/or debilitated patients and in Asian patients. Routine use of loading doses is not recommended as this practice may increase hemorrhagic and other complications and does not offer more rapid protection against clot formation.
Individualize the duration of therapy for each patient. In general, anticoagulant therapy should be continued until the danger of thrombosis and embolism has passed.
Dosing Recommendations without Consideration of Genotype
If the patient’s CYP2C9 and VKORC1 genotypes are not known, the initial dose of warfarin sodium tablets is usually 2 to 5 mg once daily. Determine each patient’s dosing needs by close monitoring of the INR response and consideration of the indication being treated. Typical maintenance doses are 2 to 10 mg once daily.
Dosing Recommendations with Consideration of Genotype
Table 1 displays three ranges of expected maintenance warfarin sodium tablets doses observed in subgroups of patients having different combinations of CYP2C9 and VKORC1 gene variants. If the patient’s CYP2C9 and/or VKORC1 genotype are known, consider these ranges in choosing the initial dose. Patients with CYP2C9 *1/*3, *2/*2, *2/*3, and *3/*3 may require more prolonged time (>2 to 4 weeks) to achieve maximum INR effect for a given dosage regimen than patients without these CYP variants.
Please review the complete therapeutic recommendations that are located here: (1)
2017 Summary of recommendations from the Dutch Pharmacogenetics Working Group (DPWG) of the Royal Dutch Association for the Advancement of Pharmacy (KNMP)
VKORC1 CT: warfarin
NO action is required for this gene-drug interaction.
The genetic variation results in a reduction in the required dose and an increase in the risk of excessively severe inhibition of blood clotting during the first month of the treatment. However, the effect is small and CT is also the most common genotype, meaning that the standard treatment will primarily be based on patients with this genotype.
VKORC1 TT: warfarin
The genetic variation results in increased sensitivity to warfarin. This results in an increase in the risk of excessively severe inhibition of blood clotting (INR >4) during the first month of the treatment.
Recommendation:
- 1.
use 60% of the standard initial dose
The genotype-specific initial dose and maintenance dose can be calculated using an algorithm, as used in EU-PACT: see https://www.knmp.nl/patientenzorg/medicatiebewaking/farmacogenetica.
From day 6 on the standard algorithm without genotype information can be used to calculate the dose.
CYP2C9 IM: warfarin
This gene variation reduces the conversion of warfarin to inactive metabolites. This can increase the risk of bleeding.
Recommendation:
- 1.
use 65% of the standard initial dose
The genotype-specific initial dose and maintenance dose can be calculated using an algorithm. Algorithms for Caucasian patients usually contain only the \*2 and \*3 allele. If the activity of the reduced-activity alleles is comparable to the activity of \*2 or \*3, then the algorithm can be completed as if \*1/\*2 or \*1/\*3 is present. See https://www.knmp.nl/patientenzorg/medicatiebewaking/farmacogenetica for Excel files containing calculation modules for oral and equivalent intravenous doses. From day 6 on the standard algorithm without genotype information can be used to calculate the dose.
Modified dose algorithms have been developed for patients of African or (East) Asian heritage.
CYP2C9 PM: warfarin
This gene variation reduces the conversion of warfarin to inactive metabolites. This can increase the risk of bleeding.
Recommendation:
- 1.
use 20% of the standard initial dose
The genotype-specific initial dose and maintenance dose can be calculated using an algorithm. Algorithms for Caucasian patients usually contain only the \*2 and \*3 allele. If the activity of the reduced-activity alleles is comparable to the activity of \*2 or \*3, then the algorithm can be completed as if \*2 or \*3 is present. See https://www.knmp.nl/patientenzorg/medicatiebewaking/farmacogenetica for Excel files containing calculation modules for oral and equivalent intravenous doses. From day 6 on the standard algorithm without genotype information can be used to calculate the dose.
Modified dose algorithms have been developed for patients of African or (East) Asian heritage.
CYP2C9*1/*2: warfarin
NO action is required for this gene-drug interaction.
Genetic variation may lead to a decrease in the required maintenance dose. However, there is insufficient evidence that this causes problems when therapy is initiated as usual.
Please review the complete therapeutic recommendations located here: ( 2, 3 )
2017 Statement from the Clinical Pharmacogenetics Implementation Consortium (CPIC)
Non-African ancestry recommendation
In patients who self-identify as non-African ancestry, the recommendation is to:
- 1.
Calculate warfarin dosing using a published pharmacogenetic algorithm, including genotype information for VKORC1-1639G>A and CYP2C9*2 and *3. In individuals with genotypes associated with CYP2C9 poor metabolism (e.g., CYP2C9 *2/*3, *3/*3) or both increased sensitivity (VKORC1-1639 A/A) and CYP2C9 poor metabolism, an alternative oral anticoagulant might be considered. The bulk of the literature informing these recommendations is in European and Asian ancestry populations, but consistent data exist for other non-African populations. These recommendations are graded as STRONG.
- 2.
If a loading dose is to be utilized, the EU-PACT loading dose algorithm that incorporates genetic information could be used. This recommendation is OPTIONAL.
- 3.
While CYP2C9*5, *6, *8, or *11 variant alleles are commonly referred to as African-specific alleles, they can occur among individuals who do not identify as, or know of their, African ancestry. If these variant alleles are detected, decrease calculated dose by 15–30% per variant allele or consider an alternative agent. Larger dose reductions might be needed in patients homozygous for variant alleles (i.e., 20–40%, e.g., CYP2C9*2/*5). This recommendation is graded as OPTIONAL.
- 4.
If the CYP4F2*3 (i.e., c.1297A, p.433Met) allele is also detected, increase the dose by 5–10%. This recommendation is also considered OPTIONAL.
- 5.
The data do not suggest an association between rs12777823 genotype and warfarin dose in non-African Americans, thus rs12777823 should not be considered in these individuals (even if available).
African ancestry recommendation
In patients of African ancestry, CYP2C9*5, *6, *8, *11 are important for warfarin dosing. If these genotypes are not available, warfarin should be dosed clinically without consideration for genotype. If CYP2C9*5, *6, *8, and *11 are known, then the recommendation is to:
- 1.
Calculate warfarin dose using a validated pharmacogenetic algorithm, including genotype information for VKORC1 c.-1639G>A and CYP2C9*2 and *3;
- 2.
If the individual carries a CYP2C9*5, *6, *8, or *11 variant allele(s), decrease calculated dose by 15–30%. Larger dose reductions might be needed in patients who carry two variant alleles (e.g., CYP2C9*5/*6) (i.e., 20–40% dose reduction).
- 3.
In addition, rs12777823 is associated with warfarin dosing in African Americans (mainly originating from West Africa). Thus, in African Americans a dose reduction of 10–25% in those with rs12777823 A/G or A/A genotype is recommended. These recommendations are considered MODERATE.
In individuals with genotypes that predict CYP2C9 poor metabolism or who have increased warfarin sensitivity (VKORC1 c.-1639 A/A) and CYP2C9 poor metabolism, an alternative oral anticoagulant should be considered (see Supplemental Material for definitions of strength of recommendations). As noted above, for non-African ancestry, if a loading dose is to be used, the EU-PACT algorithm that incorporates genetic information could be used to calculate loading dose. This recommendation is OPTIONAL. The data do not support an impact on clinical phenotype for CYP4F2 on warfarin dosing in those of African ancestry and so no recommendation is made for use of CYP4F2 genotype data in blacks.
Please review the complete therapeutic recommendations, including recommendations for pediatric patients, located here: (4).
Nomenclature
Nomenclature for Selected CYP2C9 Alleles
| Common allele name | Alternative names | HGVS reference sequence | dbSNP reference identifier for allele location | |
|---|---|---|---|---|
| Coding | Protein | |||
| CYP2C9*2 | 430C>T Arg144Cys | NM_000771.3:c.430C>T | NP_000762.2:p.Arg144Cys | rs1799853 |
| CYP2C9*3 | 1075A>C Ile359Leu | NM_000771.3:c.1075A>C | NP_000762.2:p.Ile359Leu | rs1057910 |
| CYP2C9*5 | 1080C>G Asp360Glu | NM_000771.3:c.1080C>G | NP_000762.2:p.Asp360Glu | rs28371686 |
| CYP2C9*6 | 817delA Lys273Argfs | NM_000771.3:c.817delA | NP_000762.2:p.Lys273Argfs | rs9332131 |
| CYP2C9*8 | 449G>A Arg150His | NM_000771.3:c.449G>A | NP_000762.2:p.Arg150His | rs7900194 |
| CYP2C9*11 | 1003C>T Arg335Trp | NM_000771.3:c.1003C>T | NP_000762.2:p.Arg335Trp | rs28371685 |
HGVS - Human Genome Variation Society, dbSNP - Single Nucleotide Polymorphism Database
Nomenclature for Selected VKORC1 Alleles
| Common allele name | Alternative names | HGVS reference sequence | dbSNP reference identifier for allele location | |
|---|---|---|---|---|
| Coding | Protein | |||
| -1639G>A | 1173C>T | NM_024006.4:c.-1639G>A | Not applicable - variant occurs in a non-coding region | rs9923231 |
HGVS - Human Genome Variation Society, dbSNP - Single Nucleotide Polymorphism Database
Nomenclature for Selected CYP4F2 Alleles
| Common allele name | Alternative names | HGVS reference sequence | dbSNP reference identifier for allele location | |
|---|---|---|---|---|
| Coding | Protein | |||
| CYP4F2*3 | 1297G>A Val433Met | NM_001082.4:c.1297G>A | NP_001073.3:p.Val433Met | rs2108622 |
HGVS - Human Genome Variation Society, dbSNP - Single Nucleotide Polymorphism Database
Nomenclature for rs12777823
| HGVS reference sequence | dbSNP reference identifier for allele location |
|---|---|
| NC_000010.11:g.94645745G>A (GRCh38) NC_000010.10:g.96405502G>A (GRCh37) | rs12777823 |
HGVS - Human Genome Variation Society, dbSNP - Single Nucleotide Polymorphism Database
Pharmacogenetic Allele Nomenclature: International Workgroup Recommendations for Test Result Reporting (43).
Guidelines for the description and nomenclature of gene variations are available from the Human Genome Variation Society (HGVS).
Nomenclature for Cytochrome P450 enzymes is available from Pharmacogene Variation (PharmVar) Consortium.
Acknowledgments
The author would like to thank the following experts for reviewing this summary:
Saeed Alzghari, MS, MBA (HOM), PharmD, BCPS, Director of Clinical Pharmacy, Gulfstream Genomics, Dallas, TX, USA; Bernard Esquivel, MD, PhD, MHA, President of the Latin American Association for Personalized Medicine, Mexico City, Mexico; Christine M. Formea, PharmD, BCPS, FCCP, FASHP, Pharmacogenomics Medication Therapy Management Pharmacist and Assistant Professor of Pharmacy, Mayo Clinic College of Medicine and Science, Rochester, MN, USA; Inge Holsappel, Pharmacist at the Royal Dutch Pharmacists Association (KNMP), the Hague, the Netherlands (for reviewing the information regarding the guidelines of the DPWG); George P. Patrinos, Associate Professor of Pharmacogenomics and Pharmaceutical Biotechnology, Department of Pharmacy, University of Patras, Patras, Greece; Chakradhara Rao S Uppugunduri, Maître-assistant at the CANSEARCH Laboratory, Department of Pediatrics, University of Geneva, Geneva, Switzerland.
Second edition (2016):
The author would like to thank Brian F. Gage, MD, MSC, Professor of Medicine, Washington University, St. Louis, MO, USA; and Sol Schulman, MD, Clinical Fellow in Medicine, Division of Hemostasis and Thrombosis, Department of Medicine, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, for reviewing this summary.
First edition (2012):
The Pharmacogenomics Knowledgebase: http://www.pharmgkb.org
The Clinical Pharmacogenetics Implementation Consortium: http://www.pharmgkb.org/page/cpic
Version History
To view the 2016 version of this summary (Created: June 8, 2016) please click here.
References
- 1.
- WARFARIN SODIUM- warfarin tablet [package insert]. Bridgewater, NJ; March 31, 2017. Available from: https://dailymed
.nlm .nih.gov/dailymed/drugInfo .cfm?setid=541c9a70-adaf-4ef3-94ba-ad4e70dfa057. - 2.
- Royal Dutch Pharmacists Association (KNMP). Dutch Pharmacogenetics Working Group (DPWG). Pharmacogenetic Guidelines [Internet]. Netherlands. Warfarin – CYP2C9 [Cited May 2017]. Available from: http://kennisbank
.knmp.nl [Access is restricted to KNMP membership.] - 3.
- Royal Dutch Pharmacists Association (KNMP). Dutch Pharmacogenetics Working Group (DPWG). Pharmacogenetic Guidelines [Internet]. Netherlands. Warfarin – VKORC1 [Cited May 2017]. Available from: http://kennisbank
.knmp.nl [Access is restricted to KNMP membership.] - 4.
- Johnson J.A., Caudle K.E., Gong L., Whirl-Carrillo M., et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin Pharmacol Ther. 2017 Feb 15;102(3):397–404. [PMC free article: PMC5546947] [PubMed: 28198005]
- 5.
- Basu S., Aggarwal P., Kakani N., Kumar A. Low-dose maternal warfarin intake resulting in fetal warfarin syndrome: In search for a safe anticoagulant regimen during pregnancy. Birth Defects Res A Clin Mol Teratol. 2016 Feb;106(2):142–7. [PubMed: 26389802]
- 6.
- UpToDate. Use of anticoagulants during pregnancy and postpartum [Cited October 10, 2017]. Available from: https://www
.uptodate .com/contents/use-of-anticoagulants-during-pregnancy-and-postpartum. - 7.
- PharmGKB [Internet]. Palo Alto (CA): Stanford University. Warfarin Pathway Pharmacodynamics: Simplified diagram of the target of warfarin action and downstream genes and effects [Cited 2012 Feb 24]. Available from: http://www
.pharmgkb.org /pathway/PA145011114. - 8.
- PharmGKB [Internet]. Palo Alto (CA): Stanford University. Warfarin Pathway Pharmacokinetics: Representation of the candidate genes involved in transport, metabolism and clearance of warfarin [Cited 2012 Feb 24]. Available from: http://www
.pharmgkb.org /pathway/PA145011113. - 9.
- PharmGKB [Internet]. Palo Alto (CA): Stanford University. Annotation of CPIC Guideline for warfarin and CYP2C9, CYP4F2, VKORC1: Look up your warfarin dosing guideline using the IWPC Pharmacogenetic Dosing Algorithm. [Cited 2018 January 12]. Available from: https://www
.pharmgkb .org/guideline/PA166104949. - 10.
- PharmGKB [Internet]. Palo Alto (CA): Stanford University. VIP Variant in VKORC1 [Cited 2012 Feb 24]. Available from: http://www
.pharmgkb.org /rsid/rs9923231#tabview=tab2. - 11.
- Geisen C., Watzka M., Sittinger K., Steffens M., et al. VKORC1 haplotypes and their impact on the inter-individual and inter-ethnical variability of oral anticoagulation. Thrombosis and haemostasis. 2005 Oct;94(4):773–9. [PubMed: 16270629]
- 12.
- Obayashi K., Nakamura K., Kawana J., Ogata H., et al. VKORC1 gene variations are the major contributors of variation in warfarin dose in Japanese patients. Clinical pharmacology and therapeutics. 2006 Aug;80(2):169–78. [PubMed: 16890578]
- 13.
- Ross K.A., Bigham A.W., Edwards M., Gozdzik A., et al. Worldwide allele frequency distribution of four polymorphisms associated with warfarin dose requirements. Journal of human genetics. 2010 Sep;55(9):582–9. [PubMed: 20555338]
- 14.
- Loebstein R., Dvoskin I., Halkin H., Vecsler M., et al. A coding VKORC1 Asp36Tyr polymorphism predisposes to warfarin resistance. Blood. 2007 Mar 15;109(6):2477–80. [PubMed: 17110455]
- 15.
- Rost S., Fregin A., Ivaskevicius V., Conzelmann E., et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004 Feb 5;427(6974):537–41. [PubMed: 14765194]
- 16.
- Sistonen J., Fuselli S., Palo J.U., Chauhan N., et al. Pharmacogenetic variation at CYP2C9, CYP2C19, and CYP2D6 at global and microgeographic scales. Pharmacogenetics and genomics. 2009 Feb;19(2):170–9. [PubMed: 19151603]
- 17.
- Solus J.F., Arietta B.J., Harris J.R., Sexton D.P., et al. Genetic variation in eleven phase I drug metabolism genes in an ethnically diverse population. Pharmacogenomics. 2004 Oct;5(7):895–931. [PubMed: 15469410]
- 18.
- Lee C.R., Goldstein J.A., Pieper J.A. Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002 Apr;12(3):251–63. [PubMed: 11927841]
- 19.
- PharmGKB [Internet]. Palo Alto (CA): Stanford University. Haplotype CYP2C9*2 [Cited 2012 Feb 22]. Available from: http://www
.pharmgkb.org /haplotype/PA165816543. - 20.
- PharmGKB [Internet]. Palo Alto (CA): Stanford University. Haplotype CYP2C9*3 [Cited 2012 Feb 22]. Available from: http://www
.pharmgkb.org /haplotype/PA165816544. - 21.
- Higashi M.K., Veenstra D.L., Kondo L.M., Wittkowsky A.K., et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002 Apr 3;287(13):1690–8. [PubMed: 11926893]
- 22.
- Aithal G.P., Day C.P., Kesteven P.J., Daly A.K. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999 Feb 27;353(9154):717–9. [PubMed: 10073515]
- 23.
- Limdi N.A., McGwin G., Goldstein J.A., Beasley T.M., et al. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clinical pharmacology and therapeutics. 2008 Feb;83(2):312–21. [PMC free article: PMC2683398] [PubMed: 17653141]
- 24.
- Lindh J.D., Holm L., Andersson M.L., Rane A. Influence of CYP2C9 genotype on warfarin dose requirements--a systematic review and meta-analysis. European journal of clinical pharmacology. 2009 Apr;65(4):365–75. [PubMed: 19031075]
- 25.
- Mizzi C., Dalabira E., Kumuthini J., Dzimiri N., et al. A European Spectrum of Pharmacogenomic Biomarkers: Implications for Clinical Pharmacogenomics. PLoS One. 2016;11(9):e0162866. [PMC free article: PMC5026342] [PubMed: 27636550]
- 26.
- Johnson J.A., Gong L., Whirl-Carrillo M., Gage B.F., et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clinical pharmacology and therapeutics. 2011 Oct;90(4):625–9. [PMC free article: PMC3187550] [PubMed: 21900891]
- 27.
- McDonald M.G., Rieder M.J., Nakano M., Hsia C.K., et al. CYP4F2 is a vitamin K1 oxidase: An explanation for altered warfarin dose in carriers of the V433M variant. Mol Pharmacol. 2009 Jun;75(6):1337–46. [PMC free article: PMC2684883] [PubMed: 19297519]
- 28.
- Caldwell M.D., Awad T., Johnson J.A., Gage B.F., et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008 Apr 15;111(8):4106–12. [PMC free article: PMC2288721] [PubMed: 18250228]
- 29.
- Danese E., Montagnana M., Johnson J.A., Rettie A.E., et al. Impact of the CYP4F2 p.V433M polymorphism on coumarin dose requirement: systematic review and meta-analysis. Clin Pharmacol Ther. 2012 Dec;92(6):746–56. [PMC free article: PMC3731755] [PubMed: 23132553]
- 30.
- Liang R., Wang C., Zhao H., Huang J., et al. Influence of CYP4F2 genotype on warfarin dose requirement-a systematic review and meta-analysis. Thromb Res. 2012 Jul;130(1):38–44. [PubMed: 22192158]
- 31.
- Perera M.A., Cavallari L.H., Limdi N.A., Gamazon E.R., et al. Genetic variants associated with warfarin dose in African-American individuals: a genome-wide association study. Lancet. 2013 Aug 31;382(9894):790–6. [PMC free article: PMC3759580] [PubMed: 23755828]
- 32.
- Verhoef T.I., Redekop W.K., Daly A.K., van Schie R.M., et al. Pharmacogenetic-guided dosing of coumarin anticoagulants: algorithms for warfarin, acenocoumarol and phenprocoumon. Br J Clin Pharmacol. 2014 Apr;77(4):626–41. [PMC free article: PMC3971980] [PubMed: 23919835]
- 33.
- Nagai R., Ohara M., Cavallari L.H., Drozda K., et al. Factors influencing pharmacokinetics of warfarin in African-Americans: implications for pharmacogenetic dosing algorithms. Pharmacogenomics. 2015;16(3):217–25. [PMC free article: PMC7347085] [PubMed: 25712185]
- 34.
- Ramirez A.H., Shi Y., Schildcrout J.S., Delaney J.T., et al. Predicting warfarin dosage in European-Americans and African-Americans using DNA samples linked to an electronic health record. Pharmacogenomics. 2012 Mar;13(4):407–18. [PMC free article: PMC3361510] [PubMed: 22329724]
- 35.
- Verhoef T.I., Ragia G., de Boer A., Barallon R., et al. A randomized trial of genotype-guided dosing of acenocoumarol and phenprocoumon. N Engl J Med. 2013 Dec 12;369(24):2304–12. [PubMed: 24251360]
- 36.
- Stergiopoulos K., Brown D.L. Genotype-guided vs clinical dosing of warfarin and its analogues: meta-analysis of randomized clinical trials. JAMA Intern Med. 2014 Aug;174(8):1330–8. [PubMed: 24935087]
- 37.
- Kimmel S.E., French B., Kasner S.E., Johnson J.A., et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013 Dec 12;369(24):2283–93. [PMC free article: PMC3942158] [PubMed: 24251361]
- 38.
- Furie B. Do pharmacogenetics have a role in the dosing of vitamin K antagonists? N Engl J Med. 2013 Dec 12;369(24):2345–6. [PubMed: 24251364]
- 39.
- Finkelman B.S., French B., Bershaw L., Kimmel S.E. Factors affecting time to maintenance dose in patients initiating warfarin. Pharmacoepidemiol Drug Saf. 2015 Mar;24(3):228–36. [PMC free article: PMC4570031] [PubMed: 25504915]
- 40.
- Belley-Cote E.P., Hanif H., D'Aragon F., Eikelboom J.W., et al. Genotype-guided versus standard vitamin K antagonist dosing algorithms in patients initiating anticoagulation. A systematic review and meta-analysis. Thromb Haemost. 2015 Oct;114(4):768–77. [PubMed: 26158747]
- 41.
- Verschuren J.J., Trompet S., Wessels J.A., Guchelaar H.J., et al. A systematic review on pharmacogenetics in cardiovascular disease: is it ready for clinical application? Eur Heart J. 2012 Jan;33(2):165–75. [PubMed: 21804109]
- 42.
- Gage B.F., Bass A.R., Lin H., Woller S.C., et al. Effect of Genotype-Guided Warfarin Dosing on Clinical Events and Anticoagulation Control Among Patients Undergoing Hip or Knee Arthroplasty: The GIFT Randomized Clinical Trial. JAMA. 2017 Sep 26;318(12):1115–1124. [PMC free article: PMC5818817] [PubMed: 28973620]
- 43.
- Kalman L.V., Agundez J., Appell M.L., Black J.L., et al. Pharmacogenetic allele nomenclature: International workgroup recommendations for test result reporting. Clin Pharmacol Ther. 2016 Feb;99(2):172–85. [PMC free article: PMC4724253] [PubMed: 26479518]
Footnotes
- 1
The FDA labels specific drug formulations. We have substituted the generic names for any drug labels in this excerpt. The FDA may not have labeled all formulations containing the generic drug. Certain terms, genes and genetic variants may be corrected in accordance to nomenclature standards, where necessary. We have given the full name of abbreviations, shown in square brackets, where necessary.
- Celecoxib Therapy and CYP2C9 Genotype
- Dronabinol Therapy and CYP2C9 Genotype
- Flurbiprofen Therapy and CYP2C9 Genotype
- Lesinurad Therapy and CYP2C9 Genotype
- Phenytoin Therapy and HLA-B*15:02 and CYP2C9 Genotype
- Piroxicam Therapy and CYP2C9 Genotype
- Prasugrel Therapy and CYP Genotype
- Siponimod Therapy and CYP2C9 Genotype
- Validation of a proposed warfarin dosing algorithm based on the genetic make-up of Egyptian patients.[Mol Diagn Ther. 2013]Validation of a proposed warfarin dosing algorithm based on the genetic make-up of Egyptian patients.Ekladious SM, Issac MS, El-Atty Sharaf SA, Abou-Youssef HS. Mol Diagn Ther. 2013 Dec; 17(6):381-90.
- VKORC1 and CYP2C9 genotypes are predictors of warfarin-related outcomes in children.[Pediatr Blood Cancer. 2014]VKORC1 and CYP2C9 genotypes are predictors of warfarin-related outcomes in children.Shaw K, Amstutz U, Hildebrand C, Rassekh SR, Hosking M, Neville K, Leeder JS, Hayden MR, Ross CJ, Carleton BC. Pediatr Blood Cancer. 2014 Jun; 61(6):1055-62. Epub 2014 Jan 29.
- Influence of CYP2C9, VKORC1, and CYP4F2 polymorphisms on the pharmacodynamic parameters of warfarin: a cross-sectional study.[Pharmacol Rep. 2021]Influence of CYP2C9, VKORC1, and CYP4F2 polymorphisms on the pharmacodynamic parameters of warfarin: a cross-sectional study.Sridharan K, Al Banna R, Malalla Z, Husain A, Sater M, Jassim G, Otoom S. Pharmacol Rep. 2021 Oct; 73(5):1405-1417. Epub 2021 Apr 3.
- Review Reviews of Selected Pharmacogenetic Tests for Non-Cancer and Cancer Conditions[ 2008]Review Reviews of Selected Pharmacogenetic Tests for Non-Cancer and Cancer ConditionsRaman G, Trikalinos TA, Zintzaras E, Kitsios G, Ziogas D, Ip S, Lau J. 2008 Nov 12
- Review Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing.[Clin Pharmacol Ther. 2011]Review Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing.Johnson JA, Gong L, Whirl-Carrillo M, Gage BF, Scott SA, Stein CM, Anderson JL, Kimmel SE, Lee MT, Pirmohamed M, et al. Clin Pharmacol Ther. 2011 Oct; 90(4):625-9. Epub 2011 Sep 7.
- Warfarin Therapy and VKORC1 and CYP Genotype - Medical Genetics SummariesWarfarin Therapy and VKORC1 and CYP Genotype - Medical Genetics Summaries
Your browsing activity is empty.
Activity recording is turned off.
See more...
