NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
McEntyre J, Ostell J, editors. The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2002-.
This publication is provided for historical reference only and the information may be out of date.
Summary
One of the most intensely studied regions of the human genome is the Major Histocompatibility Complex (MHC), a group of genes that occupies approximately 4–6 megabases on the short arm of chromosome 6. The MHC genes, known in humans as Human Leukocyte Antigen (HLA) genes, are highly polymorphic and encode molecules involved in the immune response. The MHC database, dbMHC, was designed to provide a neutral platform where the HLA community can submit, edit, view, and exchange MHC data. It currently consists of an interactive Alignment Viewer for HLA and related genes, an MHC microsatellite database (dbMHCms), a sequence interpretation site for Sequencing Based Typing (SBT), and a Primer/Probe database. dbMHC staff are in the process of creating a new database that will house a wide variety of HLA data including:
- Detailed single nucleotide polymorphism (SNP) mapping data of the HLA region
- KIR gene analysis data
- HLA diversity/anthropology data
- Multigene haplotype data
- HLA/disease association data
- Peptide binding prediction data
The MHC database is fully integrated with other NCBI resources, as well as with the International Histocompatibility Working Group (IHWG) Web site, and provides links to the IMmunoGeneTics HLA (IMGT/HLA) database.
This chapter provides a detailed description of current dbMHC resources and reviews dbMHC content and data computation protocols.
Introduction
The most important known function of HLA genes is the presentation of processed peptides (antigens) to T cells, although there are many genes in the HLA region yet to be characterized, and work continues to locate and analyze new genes in and around the MHC (1–4). The vast amount of work required to elucidate the MHC led to a series of International Histocompatibility Workshop/Conferences (IHWCs). These conferences were the genesis for the IHWG, a group of researchers actively involved in projects that lead to shared data resources for the HLA community (5–16). dbMHC is a permanent public archive that was conceived by the IHWG and is implemented and hosted by NCBI. This database is intended to be expandable; if you have ideas for additional resources, please send an email to vog.hin.mln.ibcn@grebmleh.
The staff of the MHC database are currently accepting online submissions for the Primer/Probe/Mix component of dbMHC. Submissions can include typing data from any of the following: Sequence Specific Oligonucleotides (SSOs), Sequence Specific Primers (SSPs), SSO and/or SSP mixes, HLA typing kits, and Sequencing Based Typing (SBT). dbMHC allows submitters to edit their submissions online at any time.
dbMHC Resources
Box 1 contains information on setting up accounts for using dbMHC.
Box 1
dbMHC guests and dbMHC accounts.
Alignment Viewer
The Alignment Viewer is designed to display pre-compiled allele sequence alignments in an aligned or FASTA format for selected loci. It offers an interactive display of the alignment, where users can select alleles, highlight SNPs within the alignment, and change between a Codon display and a Decade display (blocks of 10 nucleotides). Users can also switch the type of display, from one where the sequence is completely written out to one where only the differences between selected sequences and a pre-selected reference sequence are seen. All sequences displayed in the Alignment Viewer can be downloaded and formatted as alignment, FASTA, or XML. If the Alignment option is selected, it is up to the user to define how the alignment should be organized. The Alignment option allows the user to download the entire alignment, or just a section of it, and also allows the user to specify how the sequence will be displayed (e.g., in groups of 10 nucleotides or amino acids).
dbMHC can be used as a tool to help design primers/probes, to evaluate the reactivity patterns of potential probes, and to evaluate the polymorphism content of a particular gene.
MHC Microsatellite Database (dbMHCms)
dbMHCms contains many of the known microsatellites across the HLA region and is designed to search for descriptive information, when available, about these highly heterozygotic short tandem repeats (STRs). The MHC microsatellite information that users will be able to extract using dbMHCms includes:
- Physical location
- Number and length of known alleles
- Allelic motifs
- Informativity
- Heterozygosity
- Primer sequences
Users will find the markers shown on the dbMHCms Web page useful because they provide evidence for genetic linkage and/or association of a genomic region with disease susceptibility. This resource was developed by NCBI in collaboration with A. Foissac, M. Salhi, and Anne Cambon-Thomsen, who provided the original data on MHC microsatellites in a series of updates (17–19). dbMHCms will be expanded to include the STR markers used in the 13th IHWG workshop.
Sequencing Based Typing (SBT) Interface
The SBT interface is designed to analyze sequence information provided by users and provides an interpretation of the sequence that includes potential allele assignments and division of the sequence into exons and introns. Sequence can be submitted to the SBT interface by copying and pasting or by uploading from a file, either as haploid strands or heterozygote sequence. The current format restricts input to text or FASTA-formatted sequence.
Submitted sequences are aligned to reference locus sequences located in the dbMHC allele database based on a user-defined degree of nucleotide mismatch. After a brief analysis, the SBT interface will display exons, introns, and untranslated regions for each sequence. Allele assignments are listed according to matching order. Mismatched nucleotide positions are listed separately (in the lower frame of the SBT interface) after selecting Alignment. Note that the SBT interface analyzes one or more sequences for a single locus. If sequences from multiple loci that have identical group-specific amplifications are submitted, the SBT interface must be used to analyze sequences from only one locus at a time.
Primer/Probe Database
This database is designed to provide a comprehensive and standardized characterization of individual typing primers and probes, as well as primer and/or probe mixes, and encompasses the following technologies:
- 1.
Probe hybridization using Sequence Specific Oligonucleotides (SSOs).
- 2.
DNA amplification using Sequence Specific Primers (SSPs).
- 3.
Sequencing Based Typing (SBT) protocols. The Primer/Probe database can be used as a reference resource for probe design and is intended to provide necessary tools for the exchange of DNA typing data based on the above technologies. This database is composed of a Primer/ Probe Interface and a Typing Kit Interface. Each interface has a series of interactive frames that allow the user to access different database functions.
Primer/Probe Interface
The Primer/Probe Interface provides information on individual reagents used for the typing of MHC or MHC-related loci. Users can access this information through the multiple functional frames within the interface. The words “primers” and “probes” represent SSOs, SSPs, SSP mixes, SSO mixes, and their combinations, which include nested PCR or SSO hybridization on group-specific amplifications. Nested PCR and group-specific amplification are both based on a primary amplification with an SSP mix.
Selection of Primers/Probes
The process of selecting a primer/probe begins by entering information in the upper frame of the Primer/Probe Interface page. Users enter search options for primers, probes, and mixes, which are grouped by type (SSO, SSP, SSO mix, and SSP mix), locus, and source (submitting institution). Primers/probes can also be selected by entering either the global or local name into the search field. Once this information is entered, the lower frame fills in with the Primer/Probe Listing. Primers/probes that are then selected by the user within the Primer/Probe Listing will be displayed in the large scroll box (in the upper frame) and can be downloaded in XML format.
Primer/Probe Listing
This function is found in the lower frame of the Primer/Probe Interface page. Results of a Primer/Probe selection are displayed here and will include a unique global ID, local name, source, locus, and type for each primer/probe result. The Global Name (global ID) of each result provides a link to the Primer/Probe View or Mix View functions, and the check box associated with each primer/probe/mix can be used to generate a list of items in the Primer/Probe Interface for subsequent download in XML format.
Box 2 contains information on global IDs.
Box 2
Global IDs.
Primer/Probe View
The Primer/Probe View is located in the upper frame of the Primer/Probe Interface and appears as a result of selecting a global ID. This view will display the entire set of data of the SSP and/or SSO primers and/or probes that the user had selected in the Primer/Probe Interface. The data displayed include:
- Local name
- Locus as specified by the submitter
- Global ID
- Date of last change (“last modified”)
- Probe orientation as specified by the submitter
- Corresponding allele sequence (Allele Seq:)
- Reagent type
- Optional filter, i.e., pre-amplification
- Version number
- Probe sequence (Probe Seq:)
- The annealing position of the 3' end of the probe as specified by the submitter
- Probe stringency
Primer/Probe View offers links that will enable a user to edit primer/probe information, change primer/probe stringency, and allow users to list alleles detected by a probe with a sequence alignment. If a list of alleles detected by a primer/probe without sequence alignment is wanted, Primer/Probe View can do this as well. Select Listed, and the results will be displayed in the Primer/Probe Allele Reactivities Listing.
Primer/Probe Edit
The primer/probe edit function allows users to enter new primers/probes into dbMHC or allows users to edit existing primer/probe data. All but the following three fields can be edited (these fields are set by dbMHC):
- Global ID
- Version number
- Date of last change
Within the primer/probe edit frame, there is an alternative input field for primer/probe sequence called Allele Sequence. If primer or probe orientation is set to “reverse”, the Allele Sequence field will reverse complement the allele sequence as displayed in the alignment to generate the appropriate primer/probe sequence.
Before submitting primer/probe data, a user should define the matching stringency of the primer or probe in the Annealing Stringency field. Once the user has selected the matching stringency, the probe can be submitted. Submission of a primer/probe triggers dbMHC to begin an allele reactivity calculation that is based on the primer/probe sequence and the selected matching stringency. The result of this calculation is a list of alleles that might be detected by the submitted primer/probe. The user will find this detectable allele list in the Reactive Allele Listing (lower frame).
Alignment, on the Primer/Probe View, opens a sequence alignment in the lower frame, which is the reactive allele alignment.
Reactive Allele Listing
The Reactive Allele Listing is located in the lower frame of the Primer/Probe Interface. It lists the alleles that might be detected by a selected primer/probe or mix (Boxes 3 and 4).
Box 3
Primer/probe reactivity.
Box 4
Submitter's reactivity scores vs. dbMHC reactivity scores.
Reactive Allele Alignment
Selecting Aligned from the Primer/Probe View results in a display of the reactive allele alignment in the lower frame of the Primer/Probe Interface. It displays the alleles that might be detected by a certain primer/probe or mix in alignment with that primer/probe sequence. By default, the first 20 alleles detected by the primer/probe or mix will be displayed. The frame for the reactive allele alignment also allows a submitter to set or edit the Allele Reactivity Score of a primer/probe in this frame. If a submitter chooses to not set a reactivity score, then dbMHC sets the submitter reactivity score to “not edited”. dbMHC will also calculate a system reactivity score for the primer/probe based on the primer/probe sequence and matching stringency. Colored option boxes indicate dbMHC's reactivity score for the primer/probe, whereas dots within the option boxes indicate the submitter's reactivity score. If the submitter's score (see Box 5) is set and differs from dbMHC's score, the dbMHC score box changes to a warning color.
Box 5
Setting allele reactivity scores.
The alignment position of the 3' end of the probe is recorded for each allele. Both the sense and the reverse alignment positions are displayed if the alignment represents an SSP mix.
Only users with permission from an account administrator will be able to edit or add additional alleles to a Reactive Allele Listing for a particular primer/probe or mix. To edit the allele reactivity list, mark the “edit reactivity list” check box in the reactive allele alignment frame.
Mix View
The Mix View function is located in the upper frame of the Primer/Probe Interface and can be accessed via the link on the global ID of the Primer/Probe Listing page.
It displays an entire set of mix data for selected SSO and SSP mixes:
- Local name
- Mix type
- Locus as specified by the submitter
- Optional filter, i.e., pre-amplification
- Global ID
- Version number
- Date of last change
- List of probes as mix elements
- Mix stringency
The Mix View function contains links to the Mix Element function, where users can change or add elements of a mix. Users will also find links to the Reactive Allele Listing, where they may list alleles detected by a selected mix or selected individual elements of a mix that do not have a sequence alignment. Finally, users will find links to the reactive allele alignment function, where they can view alleles with sequence alignments that are detected by a selected mix or selected individual elements of a mix.
Mix Edit
The Mix Edit function is located in the upper frame of the Primer/Probe Interface and allows users to enter new mixes or to edit existing mix data. Users can edit all but the following three fields (these fields are set by dbMHC):
- Global ID
- Version number
- Date of last change
Annealing stringency defines the cumulative matching stringency of the elements of the mix. For SSP mixes, both the sense and the reverse primer must react with the allele with at least the defined stringency.
Mix Element
The Mix Element function is located in the lower frame of the Primer/Probe Interface and allows users to add elements to a mix or edit existing elements of a mix. To use the Mix Element function, the user must first specify the mix to be altered in the Mix Edit function and define a source for the primers/probes that the user wants listed. The mix element function displays only SSO probes for SSO mixes and displays SSP probes from SSP mixes in a sense column and a reverse orientation column. For mixes that contain probes from different sources, users must enter the mix elements separately.
Typing Kit Interface
The Typing Kit Interface is the gateway to information on individual typing kits used for typing MHC or MHC-related loci. Users can access this information through multiple functional frames within the interface. Typing kits contained within the Typing Kit Interface consist of SSOs, SSO mixes, or SSP mixes. Elements of typing kits may interact with unamplified DNA, pre-amplified DNA, or several distinct groups of pre-amplified DNA within one locus (e.g., two different amplifications of a certain exon using a distinct variation). The elements of each typing kit will react in characteristic patterns with individual alleles. These patterns can be used to determine allelic variants or a group of allelic variants within a locus (see Box 6).
Box 6
Versions of a typing kit.
Selection of the Typing Kit
Use the Typing Kit Interface to enter search parameters for typing kits. Typing kits are grouped by:
- Type: SSO, SSP
- Locus
- Source: the submitting institution
Typing kits can also be selected by typing either the global or local name into the search field.
Search results, based on user-selected parameters, will be displayed in the Typing Kit Listing, which appears in the lower frame of the Typing Kit Interface. Typing kits selected from the Typing Kit Listing will be displayed in the large scroll box (of the upper frame). They either can be used for combined pattern interpretation of multiple kits or downloaded in XML format.
Typing Kit Listing
The Typing Kit Listing is displayed in the lower frame of the Typing Kit Interface. On the basis of the criteria selected, kits are listed with their unique global ID, local name, source, locus, and type. The global ID for each kit provides a link to the Typing Kit View (upper frame of the Typing Kit Interface). The check boxes associated with each typing kit are used to generate a list of items in the Kit Select view for subsequent interpretation of a pattern or for download in XML format.
Typing Kit View
The Typing Kit View (upper frame of the Typing Kit Interface) lists the entire set of primer/probe data for selected SSO and SSP kits. Typing kit probe data include:
- Local name
- Kit type
- Locus (with or without group-specific pre-amplifications)
- Global ID
- Version number
- Batch
If List Elements from the Typing Kit View is selected, the Typing Kit Elements list provides links to the kit components. Users will also find links to the Edit Kit Locus Groups and Elements page, where they can edit kit information, or they can use Save kit as... to access the New Typing Kit function, where they can create a new kit based on a currently displayed one (see Box 7).
Box 7
Creating a virtual kit.
Typing Kit Elements
Typing Kit Elements is displayed in the lower frame of the Typing Kit Interface and is used to display all of the elements (components) within a particular typing kit. This function will group the kit elements according to the kit locus groups and will display them as an ordered list. The global ID of each kit element serves as a link to either the Mix View or the Primer/Probe View of this element.
Edit Kit Locus: Groups and Elements
The Typing Kit Interface Locus: function (upper frame) can be considered a switchboard for assembling a typing kit for dbMHC, although kit name, batch, and type must be edited using the New Typing Kit function, accessed from New Kit on the Typing Kit Interface. The kit's global ID number, version number, and the date of last change are set by dbMHC. Users wanting to edit elements of a particular kit must select a particular locus first and then select Add locus group or Edit locus group, which will open the Edit Locus Group Elements page in the lower frame of the browser (see Box 8).
Box 8
Typing kit locus "groups".
Edit Locus Group Elements
The kit group function is located in the lower frame of the Typing Kit Interface and allows a user to add single elements to, or remove single elements from, a kit group. The kit group frame displays the elements for a particular locus selected by the user in the kit locus function. If the typing kit of interest is an SSO kit, only SSO or SSO mixes will be displayed. If the typing kit is an SSP kit, only SSP mixes will be displayed. Users can then add a primer/probe to the displayed group elements by clicking on a particular primer/probe in the left column and can remove a primer/probe from the group by selecting that element and clicking Remove.
New Typing Kit
Access to the kit edit function, New Kit, is found in the upper frame of the Typing Kit Interface and allows a user to enter or modify the name, batch, and type of typing kit. Users may change the kit type so long as the kit does not yet contain any element or group.
Typing Kit Interpretation
Access to the typing kit interpretation function, Interpret, is found on the Typing Kit Interface. It provides an online typing pattern interpretation tool. Typing kit reactivity patterns can be analyzed one at a time. Several typing kits that are used to type one sample can be analyzed in combination. An order number represents each element of a typing kit. This number provides a link to the probe reactivity alignment view of each element. The display also indicates whether, and for which elements, an allele-specific amplification has been used. Reactivity patterns can be entered either as a string or via the graphical display. Both the graphical display and the text field will update each other. The string entry accepts “1”, “+” for positive reactivity, “0”,“-” for negative reactivity, “n” for not tested, “w” for weak reactivity, and “?” for undetermined. The graphic entry symbols are green for positive reactivity, orange for weak reactivity, yellow for undetermined, white for negative reactivity, and gray for “not tested”. Users can preset the main reactivity by selecting one reactivity option and clicking on Set All. If the option cycle has been selected, repeated clicking on one reactivity field of a kit will cycle through all possible reactivities.
The reactivity string or pattern entered will be interpreted as a heterozygote allele combination. Multiple kits can be combined. Users can set the degree of tolerance, which limits the number of false-positive or false-negative reactivities per locus. Each locus is analyzed separately. Allele assignments for each locus are listed according to the number of false-positive/false-negative reactivities.
Typing Kit Pattern
Selecting Pattern from the Typing Kit Interface results in a display (in the lower frame) of a cross-tab view of allele reactivities of an individual typing kit. Alleles are listed in rows, and kit elements are listed in columns. Each element of a typing kit is represented by the respective order number. This number provides a link to the probe reactivity alignment view of each element. The display also indicates whether and for which elements an allele-specific amplification has been used.
A “+” with a green background signals a detection of an allele by a kit element, a “w” with an orange background signals a weak detection, a “?” with a yellow background signals a lack of information, and an “r” with a red background signals a rejected interaction, although originally suggested by the prediction algorithm. If a kit is designed to detect only a subset of alleles, the display will be limited to this subset (see Box 9).
Box 9
Some dbMHC/browser limitations.
Database Content
The data submitted to dbMHC are stored in a Microsoft SQL (MSSQL) relational database. Table 1 is a data dictionary that defines dbMHC's database tables or record sets. For each dbMHC table, the data dictionary provides the table name, a column name, the data type in a particular table, and a summary comment.
Table 1
Data dictionary.
The relationships between dbMHC database tables are depicted in Figure 1.
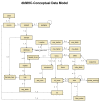
Figure 1
Physical model of the relationships among dbMHC database tables.
Primer/Probe Database
dbMHC's primer/probe database or interface is not curated. As such, the accuracy of the information presented in this database is dependent entirely upon the accuracy of the data submitted to dbMHC.
We suggest that each primer or probe submitted should be characterized by its complete sequence. In cases where submission of the complete sequence is impossible because the sequence information is considered proprietary, dbMHC offers the option to submit partial sequences. Submitted primer/probe specifications should comply with American Society of Histocompatibility and Immunogenetics (ASHI) (20) and European Federation of Immunogenetics (EFI) (21) standards for primers/probes used in histocompatibility DNA testing. If the submitted sequence contains fewer than 10 nucleotides, the submitter must also provide the position of the 3' end of the probe within the alignment of a locus. This additional information is necessary because the likelihood of a primer/probe reactivity calculation producing multiple erroneous alignment positions becomes too great if dbMHC calculates it with fewer than 10 nucleotides.
Reagent Allele Reactivity Prediction
The allele reactivity prediction tool for primers and probes available through dbMHC presents calculations based solely on sequence similarity and can therefore offer only suggestions for the possible allele reactivities of individual primers and probes. Users should not consider this tool to be 100% accurate. A reliable prediction of primer/probe allele reactivities requires complete sequence information, as well as reaction data that include annealing temperature and magnesium concentration. Because these data are not consistently available to dbMHC, we are unable to offer more than a suggestion for primer/probe allele reactivity.
Computation of Primer/Probe–Allele Interaction Stringency
dbMHC uses sequence comparisons to create a sequence interaction match or stringency grade that is compiled within a penalizing system. dbMHC's starting point for grading the interaction between a primer/probe and an allele is 100%, with each difference in sequence between the primer/probe and the allele causing a reduction in the remaining match grade by a certain percentage. The primary factors dbMHC uses to compute stringency grading include:
- Nucleotide differences
- Nucleotide position and primer/probe type
Nucleotide Differences
dbMHC divides nucleotide interactions into five categories: perfect match, high match, medium match, poor match, and no match. dbMHC defines these categories by using the purine and pyrimidine interaction between the allele and the primer/probe, as well as the number of hydrogen bonds affected during the virtual pairing of the allele with the primer/probe. See Table 2.
Table 2
Penalties for nucleotide mismatches.
Nucleotide Position and Primer/Probe Type
dbMHC uses a penalty system based on nucleotide position in its calculation of SSO/SSP interactions with different alleles. The system dbMHC uses indicates the extent to which an individual nucleotide position will affect the interaction stringency grade in a worst-case scenario, where guanines are in opposing sequence positions or cytosines are in opposing sequence positions. The maximum penalty given in the system is in the middle of an SSO and at the 3' end of an SSP, as shown in Tables 3 and 4.
Table 3
Position penalties for SSP nucleotide mismatches.
Table 4
Position penalties for SSO nucleotide mismatches.
If the initial interaction stringency grade for a particular primer/probe–allele interaction is 100% and a mismatch occurs in a position that carries a penalty of 100%, then dbMHC will reduce the interaction stringency grade to 0%. If the mismatch occurs in a position that carries a penalty of 95%, dbMHC will reduce the interaction stringency grade to 5%. A more detailed example of how dbMHC computes primer/probe interaction stringency is available online.
dbMHC's reactive allele alignment algorithm searches for sequences with a match grade above the stringency level set by the submitter of each probe. Currently, the search algorithm searches within all loci that are part of dbMHC. Within each locus, the algorithm constructs compound sequences that represent the combined polymorphic positions of alleles of the same length. Alleles with insertions or deletions are handled separately. If a primer or probe matches with a certain position within the compound, all contributing alleles are checked for that position, whether or not they match.
The reactive allele alignment algorithm will check within a specified locus for sequences that have a match grade in accordance with primer/probe specifications at a user-indicated position. The algorithm then extends SSPs toward the 5' end of the probe, to a maximum of 10 nucleotides, and extends both sides of SSOs a maximum of 15 nucleotides, observing all polymorphic positions in matching alleles. The resulting probe extension is then used by the alignment algorithm to check for cross-reactivities within other loci.
If the submitted primer/probe sequence is shorter than 10 nucleotides and the user submits a score that accepts certain alleles and rejects others, the reactive allele alignment algorithm will use this information to refine the probe extension. If the probe submitter rejects all alleles containing a unique sequence motif in the vicinity of the probe sequence, the algorithm will generate the extended probe sequence such that it does not match the unique sequence motif.
Integration with Other Resources
NCBI Links
The dbMHC provides a set of internal links to other NCBI resource homepages through either the black “quick link” bar located at the top of the dbMHC homepage or through the rotating HLA molecule located at the top left-hand corner of the dbMHC homepage. These links will take users to the following NCBI resources:
- Nucleotide
- Protein
- Molecular Modeling database (MMdb)
The dbMHC Alignment Viewer provides locus-level linkage to NCBI's LocusLink and to dbSNP at the individual SNP level.
Graphic View
The Graphic View page is a hyperlink-enabled representation of the MHC region on chromosome 6. Locus-level links from the Graphic View page include:
- dbSNP (at the SNP haplotype level)
- Map View
- LocusLink
- Nucleotide
- Protein
- Structure
- Books
The header and link column sections of dbMHC's Graphic View are the same as those on the main page. The content section of this page contains a selection box, called Choose Linked Resource, and three horizontally arranged sections, called Chromosome 6, HLA Class I, and HLA Class II. Genes listed in the HLA Class II section act as hyperlinks to the Web resource selected in Choose Linked Resource.
External Links
Currently, dbMHC provides links (from the left sidebar) to the following external sites:
The MHC haplotype project.
The International Histocompatibility Working Group (IHWG). Links to individual IHWG projects are listed in the text portion of the homepage.
The IMmunoGeneTics (IMGT)/HLA database. Links to the IMGT/HLA database are also located on the allele selection page. Users can access the allele selection page by selecting Alleles on the dbMHC Alignment Viewer. When Alleles is selected, each of the listed allele names on the allele selection page links directly to the IMGT/HLA allele-specific information page at the gene–allele level.
References
- 1.
- Bodmer J G , Marsh S G E , Albert E D , Bodmer W F , Bontrop R E , Dupont B , Erlich H A , Hansen J A , Mach B , Mayr W R , Parham P , Petersdorf E W , Sasazuki T , Schreuder G M T , Strominger J L , Svejgaard A , Terasaki P I . Nomenclature for factors of the HLA system, 1998. Tissue Antigens. 1999;53(4):407–446. [PubMed: 10321590]
- 2.
- Robinson J , Bodmer J G , Malik A , Marsh S G E . Development of the International Immunogenetics HLA Database. Hum Immunol. 1998;59(Suppl):1–17.
- 3.
- Robinson J , Marsh S G E , Bodmer J G . Eur J Immunogenet. 1999;26:75.
- 4.
- Robinson J , Bodmer J G , Marsh S G E . The American Society for Histocompatibility and Immunogenetics 25th annual meeting. New Orleans, Louisiana, USA. October 20-24, 1999. Hum Immunol. 1999;60:S1. [PubMed: 10549324]
- 5.
- Histocompatibility testing: report of a conference and workshop. Washington, DC: National Academy of Sciences - National Research Council, 1965.
- 6.
- Histocompatibility testing 1965. Copenhagen: Munksgaard, 1965.
- 7.
- Curtoni ES, Mattiuz PL, Tosi RM, eds. Histocompatibility testing 1967. Copenhagen: Munksgaard, 1967.
- 8.
- Terasaki PI, ed. Histocompatibility testing 1970. Copenhagen: Munksgaard, 1970.
- 9.
- Dausset J, Colombani J, eds. Histocompatibility testing 1972. Copenhagen: Munksgaard, 1973.
- 10.
- Kissmeyer-Nielsen F, ed. Histocompatibility testing 1975. Copenhagen: Munksgaard, 1975.
- 11.
- Bodmer WF, Batchelor JR, Bodmer JG, Festenstein H, Morris PJ, eds. Histocompatibility testing 1977. Copenhagen: Munksgaard, 1978.
- 12.
- Terasaki PI, ed. Histocompatibility testing 1980. Los Angeles: UCLA Tissue Typing Laboratory, 1980.
- 13.
- Albert ED, Baur MP, Mayr WR, eds. Histocompatibility testing 1984. Heidelberg: Springer-Verlag, 1984.
- 14.
- Dupont B, ed. Immunobiology of HLA. Vol. I. Histocompatibility testing 1987, and Vol. II. Immunogenetics and histocompatibility. New York: Springer-Verlag, 1988.
- 15.
- Tsuji K, Aizawa M, Sasazuki T, eds. HLA 1991. New York: Oxford University Press, 1992.
- 16.
- Charron, D, ed. Genetic diversity of HLA. Functional and medical implications. Paris: EDK Publishers, 1996. [PubMed: 8909580]
- 17.
- Foissac A , Salhi M , Cambon-Thomsen A . Microsatellites in the HLA region: 1999 update. Tissue Antigens. 2000;55(6):477–509. [PubMed: 10902606]
- 18.
- Foissac A , Cambon-Thomsen A . Microsatellites in the HLA region: 1998 update. Tissue Antigens. 1998;52(4):318–352. [PubMed: 9820597]
- 19.
- Foissac A , Crouau-Roy B , Faure S , Thomsen M , Cambon-Thomsen A . Microsatellites in the HLA region: an overview. Tissue Antigens. 1997;49(3 Pt 1):197–214. [PubMed: 9098926]
- 20.
- ASHI Governance. Standards for histocompatibility testing, 1998, P2.151.
- 21.
- European Federation for Immunogenetics. Modifications of the EFI standards for histocompatibility testing, 1996, P2.3100.
- 22.
- Peyret N , Seneviratne P A , Allawi H T , SantaLucia J Jr. Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A. C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. [PubMed: 10090733]
- The Major Histocompatibility Complex Database, dbMHC - The NCBI HandbookThe Major Histocompatibility Complex Database, dbMHC - The NCBI Handbook
Your browsing activity is empty.
Activity recording is turned off.
See more...

