NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
McEntyre J, Ostell J, editors. The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2002-.
This publication is provided for historical reference only and the information may be out of date.
Summary
PubMed is a database developed by the National Center for Biotechnology Information (NCBI) at the National Library of Medicine (NLM), one of the institutes of the National Institutes of Health (NIH). The database was designed to provide access to citations (with abstracts) from biomedical journals. Subsequently, a linking feature was added to provide access to full-text journal articles at Web sites of participating publishers, as well as to other related Web resources. PubMed is the bibliographic component of the NCBI's Entrez retrieval system.
Data Sources
MEDLINE®
PubMed's primary data resource is MEDLINE, the NLM's premier bibliographic database covering the fields of medicine, nursing, dentistry, veterinary medicine, the health care system, and the preclinical sciences, such as molecular biology. MEDLINE contains bibliographic citations and author abstracts from about 4,600 biomedical journals published in the United States and 70 other countries. The database contains about 12 million citations dating back to the mid-1960s. Coverage is worldwide, but most records are from English-language sources or have English abstracts.
Non-MEDLINE
In addition to MEDLINE citations, PubMed® provides access to non-MEDLINE resources, such as out-of-scope citations, citations that precede MEDLINE selection, and PubMed Central (PMC; see Chapter 9) citations. Together, these are often referred to as “PubMed-only citations.” Out-of-scope citations are primarily from general science and chemistry journals that contain life sciences articles indexed for MEDLINE, e.g., the plate tectonics or astrophysics articles from Science magazine. Publishers can also submit citations with publication dates that precede the journal's selection for MEDLINE indexing, usually because they want to create links to older content. PMC citations are taken from life sciences journals (MEDLINE or non-MEDLINE) that submit full-text articles to PMC. In addition to the incorporation of PubMed-only citations, PubMed has been enhanced recently by the incorporation of citations from the following unique databases: HealthSTAR, AIDSLINE, HISTLINE, SPACELINE, BIOETHICSLINE, and POPLINE.
In response to new approaches to electronic publishing, PubMed can now also accommodate articles published electronically in advance of being collected into an issue. We refer to these citations as "ahead of print" or "epub" citations.
Journal Selection Criteria
All content in PubMed ultimately comes from publishers of biomedical journals, and journals that are to be included in MEDLINE are subject to a selection process. The Fact Sheet on Journal Selection for Index Medicus®/MEDLINE® describes the journal selection policy, criteria, and procedures for data submission.
Electronic Data Submission
Electronic data submission benefits everyone: publishers, the NLM, and users. For the NLM, it eliminates the tremendous costs associated with entering data by hand. For publishers and users, it means that newly published data appear rapidly and accurately in PubMed. Some publishers are now making pre-publication material available before it is formally published (“ahead of print” or “epub” citations); others are publishing electronic-only journals. By close collaboration with the publisher, the citations for these publications can appear in PubMed on the same day as the article is published.
Furthermore, electronic data submission allows publishers to create links from abstracts in PubMed to the full text of the appropriate articles available on their own Web site. This can be achieved using LinkOut (Chapter 17). Both subscribers to the journals and other PubMed users can access the full text according to criteria that are determined by the publishers, increasing traffic to their sites.
Although the NLM works with many publishers directly, some publishers contract with commercial data aggregators, companies that prepare and submit the publisher's data to the NLM. Many aggregators also host publisher data on their Web sites.
Electronic Data Submission Process
All electronic data are supplied via FTP to NCBI in XML format, in accordance with the NLM's specifications (document type definition, or DTD). These specifications can be found in NLM Standard Publisher Data Format document. The document includes information on XML tag descriptions, how to handle special characters (e.g., α or β), examples of tagged records, the PubMed DTD, and a FAQ section for participating or potential data providers. Publishers or other data providers who want to submit electronic data should write to: vog.hin.mln.ibcn@rehsilbup.
NCBI staff will guide new data providers through the approval process for file submission. New providers are asked to submit test files, which are then checked for XML formatting and syntax and for bibliographic accuracy and completeness. The files are revised and resubmitted as many times as necessary until all criteria are met. Once approved, a private account is set up on our FTP site to receive new journal issues, or in the case of online publications, individual articles as they are added to the publisher's Web site. We run a file-loading script that automatically processes the files daily, Monday through Friday at approximately 9:00 a.m. (Eastern Time). The new citations are assigned a PubMed ID number (PMID), a confirmation report is sent to the provider, and the new citations usually become available in PubMed sometime after 11:00 a.m. the next day, Tuesday through Saturday.
After posting in PubMed, the citations are forwarded to NLM's Indexing Section for bibliographic data verification and for the addition of subject indexing terms from Medical Subject Headings [MeSH]. This process can take several weeks, after which time completed citations flow back into PubMed, replacing the originally submitted data.
Database Management and Hardware
PubMed is one of the NCBI databases within the relational database management system, Entrez (see Chapter 15). Entrez is a text-based search and retrieval system based on in-house software that uses an indexing system for rapid retrieval of information.
Requests for NCBI services, including PubMed, are first proxied through three load-balanced Dell PowerEdge 1650 servers, each with two central processing units. The proxy servers, in turn, load-balance requests forwarded on to the Web servers for PubMed and other NCBI services.
The PubMed Web servers comprise eight Dell PowerEdge 8450 servers. The Dell servers have eight central processing units, 8 GB of memory, and about 300 GB of disk space and run the Linux operating system.
The Web servers retrieve PubMed records from two Sybase SQL database servers, which run on Sun Enterprise 450s. To accommodate the data volume output by PubMed and other Web-based services, the NLM has a high-speed connection (OC-3, up to 155 Mbits/sec) to the Internet, as well as a 622 Mbits/sec connection (OC-12) to Internet2, the noncommercial network used by many leading research universities.
Indexing
PubMed Citation Status and Assignment of MeSH Terms
Citations in PubMed are assigned one of three citation status tags that display next to the PubMed ID (PMID) numbers on all PubMed citations. The citation status tags indicate the citation's stage in the MEDLINE indexing process. The three tags are:
[PubMed - as supplied by publisher]: This tag is displayed on citations added recently to PubMed via electronic submission from a publisher (which may or may not move on for MEDLINE MeSH indexing).
[PubMed - in process]: This tag is displayed on citations that have had the first stage of quality review to verify that the journal, date, volume, and/or issue are correct. They will be reviewed for other accurate bibliographic data at the article level (e.g., pagination, authors, article title, and abstract) and indexed, i.e., the articles will be reviewed and MeSH vocabulary will be assigned (if the subject of the article is within the scope of MEDLINE).
[PubMed - indexed for MEDLINE]: This tag is displayed on citations that have been indexed with MeSH, Publication Types, Registry Numbers, etc., and have been completely reviewed for accurate bibliographic data. This is an intellectual process of assigning controlled vocabulary terms to describe the contents of the journal article and verifying other aspects of the citation data.
Most citations that are received electronically from publishers progress through “in process” status to MEDLINE status. Those citations not indexed for MEDLINE remain tagged [PubMed - as supplied by publisher]. Citations with “in process” status proceed to MEDLINE status after MeSH terms, publication types, sequence Accession numbers, and other indexing data are added.
All records are added to PubMed Monday through Friday and become available for viewing Tuesday through Saturday. For additional information, please see the NLM Fact Sheet: What's the Difference Between MEDLINE® and PubMed®?
The Automatic Computer Indexing Process
The aim of the computer indexing process is to automatically create multiple machine-readable access points that refer to the different components of the journal citations for use when searching PubMed. The citations are loaded into PubMed from both the NLM Data Creation and Maintenance System (DCMS) and directly from journal publishers (Figure 1). Both sources are in XML.
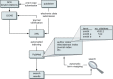
Figure 1
A schematic representation of PubMed data flow.
During the computer indexing process, the citation information is broken down into index fields such as Journal Name, Author Name, and Title/Abstract. The words in each of the fields are checked against the corresponding index (i.e., title words in a new citation are looked up in the Title/Abstract Index). If the word already exists, the PMID of the citation is listed with that index term. If the word is a new one for the Index, it is added as a new Index term, and the PMID is listed alongside it. (In the first instance that the term already exists, the new term will have only this one citation associated with it; this is how the PubMed indexes grow.)
Each PubMed citation is, therefore, associated with several indexes, and in cases similar to the Title/Abstract Index, many different index terms can refer back to a single citation. Likewise, commonly used terms will refer to thousands of citations (the term “cell”, for example, is found in the Title/Abstract of 1,092,124 citations at the time of this writing). The Field Indexes can be browsed by using PubMed's Preview/Index function.
How PubMed Queries Are Processed
Automatic Term Mapping
PubMed uses Automatic Term Mapping to process words entered in the query box by someone searching PubMed. Terms entered without a qualifier, i.e., a simple text phrase that does not specify a search field, are looked up against the following translation tables and indexes in a distinct order:
1. MeSH Translation Table
2. Journals Translation Table
3. Author Index
1. MeSH Translation Table
The MeSH Translation Table contains:
- MeSH Terms
- See-Reference mappings (also known as entry terms) for MeSH terms
- Mappings derived from the Unified Medical Language System (UMLS) that have equivalent synonyms or lexical variants in English
- Names of Substances and synonyms to the Names of Substances (now known as Supplementary Concept Substance Names)
If the search term is found in this translation table, the term will be mapped to the appropriate MeSH term, and the Indexes will be searched as both the text word entered by the user and the MeSH term:
Search term: gallstones
“Gallstones” is an entry term for the MeSH term “cholelithiasis” in the MeSH translation table.
Search translated to: “cholelithiasis” [MeSH Terms] OR gallstones [Text Word]
When a term is searched as a MeSH term, PubMed automatically searches that term plus the more specific terms underneath in the MeSH hierarchy:
Search term: breast cancer
“Breast cancer” is an entry term for the MeSH term “breast neoplasms” in the MeSH translation table.
“Breast neoplasms” has the specific headings “breast neoplasms, male”, “mammary neoplasms”, “mammary neoplasms, experimental”, and “phyllodes tumor”, all of which are also searched.
2. Journals Translation Table
If the search term(s) is not found in the MeSH Translation Table, the PubMed search algorithm then looks up the term in the Journals Translation Table, which contains the full journal title, MEDLINE abbreviation, and International Standard Serial Number (ISSN):
Search term: New England Journal of Medicine
“New England Journal of Medicine” maps to N Engl J Med.
Search translated to: “N Engl J Med” [Journal Name]
If a journal name is also a MeSH term, PubMed will search the term as both a MeSH term and as a Text Word, but not as a journal name, for a search that does not specify the “journal” field:
Search term: Cell
Search translated as: “cells” [MeSH Terms] OR cell [Text Word]
Search term: Cell [Journal]
Search translated as: “Cell” [Journal]
3. Author Index
If the phrase is not found in MeSH or the Journals Translation Table and is a word with one or two letters after it, PubMed then checks the Author Index. The author's name should be entered in the form: Last Name (space) Initials, e.g., o'malley f, smith jp, or gomez-sanchez m.
If only one initial is used, PubMed finds all names with that first initial, and if only an author's last name is entered, PubMed will search that name in All Fields. It will not default to the Author Index because an initial does not follow the last name:
Search term: o'malley f
Search translated as: o'malley fa, o'malley fb, o'malley fc, o'malley fd, o'malley f jr, etc.
Search term: o'malley
Search translated as: “o'malley” [All Fields]
A history of the NLM's author indexing policy regarding the number of authors to include in a citation is outlined in Table 1.
Table 1
History of NLM author-indexing policy.
Search Rules and Field Abbreviations
It is possible to override PubMed's Automatic Term Mapping by using search rules, syntax, and qualifying terms with search field abbreviations.
The Boolean operators AND, OR, and NOT must be entered in uppercase letters and are processed left to right. Nesting of search terms is possible by enclosing concepts in parentheses. The terms inside the set of parentheses will be processed as a unit and then incorporated into the overall strategy. Terms may be qualified using PubMed's Search Field Descriptions and Tags. Each search term should be followed by the appropriate search field tag, which indicates which field will be searched:
Search term: o'malley [au] will search only the author field. Specifying the field precludes the Automatic Term Mapping, which would result in the search o'malley[All Fields] if the field were not specified. Similarly, using the search term Cell [Journal] avoids using the MeSH Translation Table, which would interpret Cell as only a text word and MeSH term.
Using PubMed
Searching
Simple Searching
A simple search can be conducted from the PubMed homepage by entering terms in the query box and pressing Enter from the keyboard or clicking on the Go button on the screen.
If more than one term is entered in the query box, PubMed will go through the Automatic Term Mapping protocol described in the previous section, first looking for all the terms, as typed, to find an exact match. If the exact phrase is not found, PubMed clips a term off the end and repeats Automatic Term Mapping, again looking for an exact match, but this time to the abbreviated query. This continues until none of the words are found in any one of the translation tables. In this case, PubMed combines terms (with the AND Boolean operator) and applies the Automatic Term Mapping process to each individual word. PubMed ignores Stopwords, such as “about”, “of”, or “what”. People can also apply their own Boolean operators (AND, OR, NOT) to multiple search terms; the Boolean operators must be in uppercase.
Search term: vitamin c common cold
Translated as: ((“ascorbic acid” [MeSH Terms] OR vitamin c [Text Word]) AND (“common cold” [MeSH Terms] OR common cold [Text Word]))
Search term: single cell separation brain
Translated as: (((“single person” [MeSH Terms] OR single [Text Word]) AND (“cell separation” [MeSH Terms] OR cell separation [Text Word])) AND (“brain” [MeSH Terms] OR brain [Text Word]))
If a phrase of more than two terms is not found in any translation table, then the last word of the phrase is dropped, and the remainder of the phrase is sent through the entire process again. This continues, removing one word at a time, until a match is found.
If there is no match found during the Automatic Term Mapping process, the individual terms will be combined with AND and searched in All Fields.
One can see how PubMed interpreted a search by selecting Details from the Features Bar on the PubMed search pages after completing a search. For more information, see Details.
Complex Searching
There are a variety of ways that PubMed can be searched in a more sophisticated manner than simply typing search terms into the search box and selecting Go. It is possible to construct complex search strategies using Boolean operators and the various functions listed below, provided in the Features Bar:
- Limits restricts search terms to a specific search field.
- Preview/Index allows users to view and select terms from search field indexes and to preview the number of search results before displaying citations.
- History holds previous search strategies and results. The results can be combined to make new searches.
- Clipboard allows users to save or view selected citations from one search or several searches.
Additional PubMed Features
The following resources are available to facilitate effective searches:
- MeSH Database allows searching of MeSH, NLM's controlled vocabulary. Users can find MeSH terms appropriate to a search strategy, obtain information about each term, and view the terms within their hierarchical structure.
- Clinical Queries is a set of search filters developed for clinicians to retrieve clinical studies of the etiology, prognosis, diagnosis, prevention, or treatment of disorders. The Systematic Reviews feature retrieves systematic reviews and meta-analysis studies by topic.
- Journal Database allows searches of journal names, MEDLINE abbreviations, or ISSN numbers for journals that are included in the Entrez system. A list of journals with links to full text is also included.
- Single Citation Matcher is a “fill-in-the-blank” form that allows a user to find the PubMed ID (PMID) number for a single article or all citations in a given journal issue by entering partial journal citation information.
- Batch Citation Matcher allows users to find PMID numbers that correspond to their own list of citations. Publishers or other database providers who want to link directly from bibliographic references on their Web sites to entries in PubMed use this service frequently.
- Cubby is a place for users to store search strategies, LinkOut preferences, and changes to the default Document Delivery Services.
Results
PubMed retrieves and displays search results in the Summary format in the order the record was initially added to PubMed, with the most recent first. (Note that this date can differ widely from the publication date.) Citations can be viewed in several other formats and can be sorted, saved, and printed, or the full text can be ordered.
Links from PubMed
A variety of links can be found on PubMed citations including:
Related Articles, which retrieves a precalculated set of PubMed citations that are closely related to the selected article. PubMed creates this set by comparing words from the title, abstract, and MeSH terms using a word-weighted algorithm.
LinkOut, which provides links to publishers, aggregators, libraries, biological databases, sequencing centers, and other Web sites. These link to the provider's site to obtain the full text of articles or related resources, e.g., consumer health information or molecular biology database records. There may be a charge to access the text or information, depending on the policy of the provider.
Books, which provides links to textbooks so that users can explore unfamiliar concepts found in search results. In collaboration with book publishers, NCBI is adapting textbooks for the Web and linking them to PubMed. The Books link displays a facsimile of the abstract, in which some words or phrases show up as hypertext links to the corresponding terms in the books available at NCBI. Selecting a hyperlinked word or phrase takes you to a list of book entries in which the phrase is found.
NCBI databases, as well as other resources, may be available from the Links pull-down menu to the right of each citation and from the Display pull-down menu. PubMed will return only the first 500 items when using the Display pull-down menu, from which the following links are available:
- Genome – records and graphic displays of entire genomes and chromosomes for megabase-scale sequences.
- ProbeSet – gene expression data repository and online resource for the retrieval of gene expression data from any organism or artificial source.
- OMIM – directory of human genes and genetic disorders.
- SNP – dbSNP is a database of single nucleotide polymorphisms.
- Domains – The Domains database is used to identify the conserved domains present in a protein sequence.
- 3D Domains – the domains from Entrez Structure.
How to Create Hyperlinks to PubMed
The Entrez system provides three distinct ways to create Web URL links that search and retrieve items from PubMed and the molecular biology databases: (1) by using the Entrez Programming Utilities; (2) via the URL button on the Details screen; and (3) by constructing URLs by hand.
The Entrez Programming Utilities can be used to create URL links directly to all Entrez data, including PubMed citations and their link information, without using the front-end Entrez query engine. These Utilities provide a fast, efficient way to search and download citation data.
Customer Support
If you need more assistance, please contact our Customer Support services by selecting the Support Center link displayed on all PubMed pages or by sending an email to vog.hin.mln@vrestsuc. You may also contact the NLM Customer Service Desk at 1-888-346-3656 [(1-888)-FINDNLM]. Hours of operation are Monday through Friday from 8:30 a.m. to 8:45 p.m. and Saturday from 9:00 a.m. to 5:00 p.m. (Eastern Time).
Additional information is also available in the PubMed Tutorial, PubMed Training Manuals, and NLM Technical Bulletin.
- PubMed: The Bibliographic Database - The NCBI HandbookPubMed: The Bibliographic Database - The NCBI Handbook
Your browsing activity is empty.
Activity recording is turned off.
See more...

