NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
McEntyre J, Ostell J, editors. The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2002-.
This publication is provided for historical reference only and the information may be out of date.
Introduction
This chapter contains tutorials for using Map Viewer. Step-by-step instructions are provided for several common biological research problems that can be addressed by exploiting the whole-genome and positional perspectives of Map Viewer. Please be aware that the examples in these tutorials may return different results when you execute them, because the underlying data may have been updated, but we hope that the framework for obtaining, interpreting, and processing your results will be sufficiently clear if that happens. Most of the examples are for human genes, but the same logic applies to other genomes as well.
Please note that each of these tutorials is accompanied by a figure. If you are using this tutorial on the Web, we suggest that you open another browser window so that you can view the figure as you are reading the text. You are also encouraged to use Map Viewer interactively.
We welcome any suggestions that you have for improving the existing tutorials or for adding new ones.
1. How Do I Obtain the Genomic Sequence around My Gene of Interest?
There are many instances in molecular biological research when you may have only a cDNA sequence but need to have the nucleotide sequence that lies 5′ or 3′ to a gene or the introns for additional analyses. Because genomic sequence available from the public database may not have this annotation or may be so large as to make it difficult to retrieve only a region of interest, tools have been added to Map Viewer to make it easier to define, view, and download genomic sequence in multiple formats.
Direct Sequence Information via Map Viewer
From the Map Viewer homepage, select Search Homo sapiens (human) from the pull-down menu, enter FMR1 in the box labeled for, and then select Go. On the results page, four entries are returned (4 hits). FMR1 has been mapped on three different maps: Genes_cyto, Genes_seq, and Morbid. Select the Genes_seq map to see the structure of the FMR1 gene. Two links to the right of the Genes_seq map are of use in retrieving the 5′ and 3′ flanking DNA, the sv and seq links (boxed). At the top of the page, Download/View Sequence/Evidence can also be used.
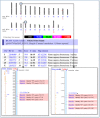
The most informative maps, for the purpose of this example, are the Contig, Component, and Genes_seq maps. Selecting the Genes_seq map will display the current gene model. To start our search for flanking DNAs, display the Contig and Component maps. To do so, select Maps & Options, and from the available maps, choose the Contig map by left-clicking with the mouse. Select ADD>>. Now add the Component map. Next, make the Gene_seq map the master map in the display by left-clicking on the Gene map under the Maps Displayed (left to right) and selecting Make Master/Move to Bottom:. Finally, select the Contig map and choose the Toggle Ruler to add a ruler ([R]) to the display. This will guide you in finding your region of interest. Select Apply. From Figure 1, we can see that FMR1 has been annotated on a finished contig, NT_011537.9 (c), and that the contig is built from two components in this region, AC016925.15 and L29074.1 [(a) and (b), respectively].
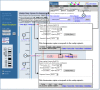
Figure 1
Making a master map.
There are two links on the Map Viewer display that are used to view and download of the region of interest: the seq link and the Download/View Sequence/Evidence link (boxed). The seq link is displayed only when the Gene_seq map is made the master map in the display. Selecting the seq link will open a window, prompting the user to enter a region to retrieve. This region can then be refined further by adjusting the position, in kilobase pairs. Selecting Display Region will change the start and stop positions on the contig, where the gene has been annotated. The region can then be displayed and saved locally in FASTA or GenBank formats.
Selecting the Download/View Sequence/Evidence link from the main display page will generate the same window, but the initial coordinates are those spanning the entire region displayed by the Map Viewer rather than the region of a particular gene, as in the case above.
Let us assume that we would like to download 5.0 kb of upstream DNA and 1.0 kb of downstream DNA. To define this region, we will need to follow the seq link, which will open a new display showing the chromosome coordinates for FMR1 and the corresponding position on the contig. To adjust the region, simply enter the amount of desired upstream DNA and downstream DNA into the two adjust by: input boxes provided, and select Change Region. Notice that the corresponding region on the contig has adjusted to reflect this change in position. Now we can either display the data in GenBank or FASTA formats and save the data to a disk.
Using the Sequence Viewer
Another tool for obtaining the desired sequence is the Sequence Viewer, available via the sv link when the Gene_seq map is the master map. The Sequence Viewer presents a graphical view of the gene within the contig. The sequence is also annotated with the coding regions, RNA and gene features, Sequence Tagged Sites (STSs), and single nucleotide polymorphisms. Blue arrows at the top and bottom of the display allow the user to navigate upstream or downstream. The Get Subsequence link (large, open arrow) at the top of the sv page allows the user to change the sequence range on the contig and will also display the reverse complement of the sequence. The specified region can then be displayed and saved locally in FASTA format.
2. If I Have Physical and/or Genetic Mapping Data, How Do I Use the Map Viewer to Find a Candidate Disease Gene in That Region?
In this example, we will use the Map Viewer to look for human candidate genes in a region. The types of queries that can be posted to the Map Viewer that will address this type of question are queries by genetic marker or STS.
Please note that Map Viewer supports queries by any named object positioned on a map so that it is possible to query by gene symbol or GenBank Accession number or any other object that might define your range of interest.
Querying by STSs
To refine our search, we will enter the names of two STSs. In the text box, enter “sWXD113 OR DXS52" on chromosome X. Select Find. We can see that these two STSs map to the distal region of the long arm of the X chromosome, Xq, by the red tick marks that appear alongside the schematic of the X chromosome. These two STS markers have been mapped on several other maps that are also represented in the results. Select the X chromosome above the red 3 to see both markers in the same display. The Map Viewer page now displays three different maps, each showing the physical location of these two markers.
The maps that are displayed include the UniG_Hs, Genes_seq, and STS maps. The UniG_Hs map shows the density of ESTs and mRNAs that align to the current assembly of the human genome. The Genes_seq map displays known and predicted genes that are annotated on the genomic contigs. The rightmost map, the STS map in this case, is termed the master map and contains descriptive information about each map element (Figure 2). The two STSs for which we are searching (GDB:192503 and DXS52) are highlighted in pink. To the right of the display is a grid indicating other maps upon which these STSs are located. The red dots (circled) to the left of each highlighted STS show the relative position of these STSs in the context of the two other maps. By default, a ruler is displayed alongside the STS map so that the region of interest can be localized further. Notice on the far left of the page that there is an area where you can enter the region that you would like to display [(a)].

Figure 2
Master Map: STS.
At the current resolution, it is not possible to view all of the information displayed on the three maps. Therefore, some adjustment will be necessary.
Identifying a Candidate Gene
Narrow the region further using the ruler adjacent to the STS map as a guide. It is not necessary to display a ruler alongside the other two maps because they are all on the same coordinate system. In instances where sequence, genetic, cytogenetic, or radiation hybrid maps are being displayed in the same view, it is advisable to display additional rulers because the different maps show the mapped element on different coordinate systems (Kbp, cM, banding position, or centiRays, respectively). Enter the range 133.0 M to147.0 M in the boxes to the left of the page, in the Region Shown [(a)]. Select Go. This is a slightly better view; however, there are many more genes in the region that can be displayed.
To see all of the genes in the region defined by the two markers, select the Data as Table View link (boxed). The table that is generated lists all 144 genes in this region in a format easily read by people or computers. The table also preserves the links to additional gene-related resources seen in the graphical display, as well as reporting other objects in your displayed region. Please note that links are also provided to make it easy for you to download reports, not only of the objects in your map display, but other objects within the region defined by your display. This feature is especially useful if you are looking for other gene markers in your region of interest.
We can also change the page length under Maps & Options to a number large enough so that all genes are displayed graphically on a single page. By default, Map Viewer will show 20 map elements on a page. When the Genes_seq map is made the master map, there will be information at the top of the page, indicating how many genes have been labeled (n = 20) and how many genes are in the region (n = 144). To see all of the candidate genes in the specified region, go to Maps & Options and change the page length to the number of genes in the region (n = 144) and then select Apply.
Interpreting Your Results
At this point, you can now browse the description of the genes that are being displayed. Each gene or locus name is hyperlinked to LocusLink, where a detailed report about the gene or locus is provided. If the gene or locus of interest has supporting EST and mRNA data, then you can select the UniGene cluster number and link to UniGene, where more detailed information is provided about this gene, including its pattern of expression. LocusLink also provides connections to BLink and thus indirectly to reports of related proteins in the protein database and to viewers of protein structure, if your protein of interest is related to a protein for which the structure is known (see also Exercise 8 in this chapter).
Other Ways to Query
The example above summarizes the approach taken when defining a region of interest by entering names of markers in the query box. Gene symbols, reference SNP names, and GenBank Accession numbers for ESTs could also be used. It should also be noted that when a chromosome is displayed, you may also submit a query using the Region Shown boxes in the bar at the left side.
3. How Can I Find and Display a Gene with the Map Viewer?
In this example, we will locate and display the human gene implicated in Fragile X syndrome using the Map Viewer. We can find the gene beginning with several types of data. Refer to Figure 3. For these examples, we assume you are starting from the human-specific Map Viewer. If you instead are starting from the homepage, please remember to select “Homo sapiens (human)” as the species.
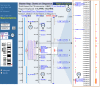
Figure 3
Location and display of the human gene implicated in Fragile X syndrome.
By Gene Symbol
If we are fortunate enough to know the official gene symbol for the Fragile X gene, FMR1, or an alternative symbol such as FRAXA, we can type the symbol into the search box at the top of the page and press the Find button. This gene has been annotated on the genome, and the gene symbol FMR1 appears on the Genes_seq, Genes_cyto, and the Morbid maps. To generate a Map Viewer display that includes all three maps, select the link to all matches.
By Linkage to a Disease
Genes that are linked to a disease in Online Mendelian Inheritance in Man (OMIM) are referenced on the Morbid map and can be found by searching with a disease name or phenotype. In our case, “fragile-X” can be used. Using this query, we pick up hits to genes related to FMR1, as well as our intended gene. Selecting the FMR1 link generates a display of the gene.
By Physical Marker
The FMR1 gene contains the STS, STS-X69962 [(b)]; therefore, we can also use the name of this STS to find it, as in the case of a gene symbol. The search yields a table of hits showing that STS-X69962 appears on the STS map. Selecting the STS link gives us a Map Viewer display of the STS we found but not of the gene we sought. To get the gene into the Map Viewer display, we can add the Genes_seq map to the display. Although FMR1 is located on the Genes_seq map rather than the STS map, because the coordinate systems used in different maps are synchronized, we can see the FMR1 gene if we ask for the Genes_seq track in the region corresponding to the hit on the STS map. To do this, we can select the Maps & Options link [(a)], highlight the Gene map from the list of available maps in the left-hand box, and select the ADD>> button to add this map to the list of displayed maps. After selecting the Apply button, the Gene map is added to the Map Viewer display. Note, however, that the view is limited to a very small 200-base pair portion of the gene. This is because we are still focused on the STS returned by our initial search. To see an expanded view, we must zoom out using the zoom control located directly over the thumbnail chromosome map in the blue sidebar [(d)]. Mousing over the control indicates that we are viewing 1/10,000th of chromosome X. We can click further up on the control to view 1/1,000th of the chromosome and see most of the FMR1 gene. The FMR1 gene is now centered in the Map Viewer display, and our STS hit is marked in red.
By Region
Often, a gene is known to reside only in a particular region. Suppose that we know only that FMR1 resides somewhere between markers DXS532 and DXS7389 [(e) and (c), respectively]. We can use a query containing a Boolean OR to force the Map Viewer to search for both markers simultaneously. In this case, a query to Map Viewer of “DXS532 OR DXS7389” generates a number of hits to various physical maps, all to a region on chromosome X, which is marked in red under the X chromosome graphic. If we select the chromosome X link under the chromosome graphic, the Map Viewer display shows marker hits on several physical maps, highlighted in red, over a fairly large sequence region from about 141 to 142 megabases. To generate a tabular listing of all the genes in this region, select the Data as Table View link to the left of the Map Viewer display. With a genetic map, such as the Genethon map, as the master map, it is also possible to define or refine the region of display using coordinates in centimorgans. In the case of FMR1, entering a range of 176–198 into the Region Shown boxes generates a display of the genes falling within that range.
By Sequence Homology
Suppose that we have the sequence of the mouse homolog of the human FMR1 gene and want to locate it on the human genome assembly. We might consult the Human/Mouse homology map at NCBI as the most direct approach for mapped genes, but let us assume that the human homolog of the FMR1 gene is unmapped. In this case, we can perform a BLAST sequence similarity search with the mouse sequence to attempt to locate the corresponding human gene. We will use the mouse Fmr1 mRNA sequence, taken from NCBI's LocusLink database (Accession number NM_002024), as our probe and follow the link to BLAST search the human genome located at the top of the Map Viewer search page; type the above Accession number into the BLAST form, and press the Search button. Such searches are extremely fast because they make use of an NCBI program called MegaBLAST, designed especially for this purpose. In the MegaBLAST results, the Genomic View button near the top of the page provides an entry point into a Map Viewer display. The MegaBLAST hits are indicated by a red mark on chromosome X, and links to hits are provided in the table at the bottom of the page, as in the searches by gene symbol or marker. In the case of any MegaBLAST search, all hits are to sequences [(f)] on the Contig map; therefore, it is the contig link (Accession number beginning with NT_) that we follow to view the hits in their genomic context. Following this link brings us to a display of the FMR1 gene, with the BLAST hits indicated as highlighted “hits” on the contig map and colored ticks on the other maps (large, expanded view).
Other Ways to Query
Please note that other resources within NCBI also support querying for genes. Consider also LocusLink, UniGene, and Entrez Nucleotide. When a record of interest has been retrieved, each of these provides links to Map Viewer.
4. How Can I Analyze a Gene Using the Map Viewer?
We will analyze the FMR1 gene. To find this gene in the human Map Viewer, enter FMR1 into the query box and select the Genes_seq map link in the table of search results.
To begin the analysis, we can select the link in the blue sidebar entitled Data as Table View to see a tabular listing of the chromosomal coordinates of the features visible in the current Map Viewer display. In the section of the table giving features on the Genes_seq map, we find that the FMR1 gene extends from 141519781 to 141558823, a span of about 40,000 base pairs. We can now return to the graphical Map Viewer display and limit our view to this region by entering this range into the Region Shown boxes [Figure 4, (a)] in the blue sidebar and pressing the Go button. The coordinate system operative in the Region Shown boxes is that of the rightmost map in the Map Viewer display, called the master map; be sure that the Genes_seq map is the master before changing the coordinates for the region shown.
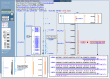
Figure 4
Set of maps.
We are now ready to select the maps to display. The maps displayed will depend on the sort of analysis intended; however, one useful set of maps includes the Genes_seq, Contig, Comp, GScan, UniG_Hs, RNA, and gbDNA maps. This set of maps can be selected for viewing using the panel invoked by the Maps & Options link (large, open arrow). A Map Viewer display of the FMR1 gene using this set of maps is given in Figure 4.
The Genes_seq Track
In Figure 4, the Genes_seq map, which displays annotated genes, is the master map and is the first one we will examine. The FMR1 gene comprises 18 exons, represented by thick lines, interspersed over about 40,000 base pairs of sequence. The gene is drawn to the right of the Genes_seq track line and therefore runs from the top of the display to the bottom and is coded on the “plus” strand in the human genome assembly. Genes located on the opposite strand run from the bottom of the display upward and are shown to the left of the Genes_seq track. The FMR1 gene is displayed in orange, which indicates that the alignment between the genomic sequence and the FMR1 transcript sequences that were used to produce the gene model was not perfect; blue alignments are of the best quality. The 3′-most exon (bottom inset) of the gene is exceptionally large and probably includes a significant untranslated region. To verify this, we can select the sv link (boxed) to the right of the Genes_seq map to invoke the Sequence Viewer, which shows the sequence of the FMR1 gene. In the Sequence Viewer, we can navigate to the display for the last exon, number 18, to see that the coding sequence ends toward the beginning of the exon and that the majority of the exon is indeed untranslated.
The RNA Track
The RNA or Transcript map shows the alignment of a single mRNA sequence to the genome, and in this case, the pattern of exons produced matches exactly that shown on the Genes_seq map. If additional splice variants are sequenced, multiple alignments will be shown on the RNA track, and the gene model given on the Genes_seq track will be a composite model made up of all the exons implied by these alignments.
The GScan Track
The GenomeScan track shows gene predictions made using GenomeScan that are independent of supporting mRNA alignments. The GenomeScan model for FMR1 is very similar to the model shown on the Genes_seq track; however, there are differences, and the alignment-based model shown on the Genes_seq track covers exons that are part of two different GenomeScan models (topmost inset). Note also that the GenomeScan model covers only the initial translated portion of the large 3′ exon of the transcript-based model (bottom inset).
The UniG_Hs Track
Both the predicted model (GScan) and the alignment-based model (Genes_seq) can be compared to the mapping of ESTs on the UniG_Hs track. The bars extending to the left of the UniG_Hs track line depict EST mapping density, whereas the lines to the right connect ESTs arising from common UniGene clusters. From the UniG_Hs track, it is clear that most of the exons arising from either of the two gene models have some EST support, and that most of the ESTs that map to these regions are members of UniGene cluster Hs.89764 (boxed, center). Selecting the Hs.89764 link leads to the FMR1 UniGene cluster.
The UniG_Mm Track
This map is comparable to the UniG_Hs track but is based instead on alignment of mouse cDNA sequences (conventional and EST). In this example, the exons suggested by the alignment of mouse cDNAs is comparable to that based on alignment of human cDNAs.
SAGE_tag
SAGE_tag provides another view of expression levels and connections to more information about the tissue of origin of the expressed sequences. The SAGE_tag map also provides a histogram of expression, and each tag is connected to a tag-specific report page.
The Contig Track
Looking across to the Contig track, we can see that the gene maps to a contig that is drawn in blue, indicating that the contig is derived from high quality, finished sequence. If we consult the Component map (Comp), we can see that the portion of the contig containing the FMR1 gene is composed of two overlapping finished sequences, also drawn in blue. Because the sequence underlying the FMR1 gene is finished, rather than draft sequence, the FMR1 sequence and structure are likely to remain stable in future human genome assemblies.
The gbDNA Track
Additional GenBank sequences that align to the genome but were not part of the assembly are shown on the gbDNA track. In this case, a number of short sequences for the individual exons of FMR1, the product of intense research on this gene, are aligned to the genome (Figure 4b).
5. How Can I Create My Own Transcript Models with the Map Viewer?
The Map Viewer displays the alignment of transcripts, such as mRNA GenBank sequences and RefSeqs, to genomic sequence and shows the positions of predicted genes, but it does not stop there. By using a utility called the ModelMaker, it is possible to combine the alignment evidence with the results of gene prediction to construct novel transcripts.
Beginning with the standard Map Viewer display for the gene FMR1, we can display the Gscan and Genes_seq maps in parallel, as shown in Figure 5. The GScan map shows gene predictions made by the GenomeScan program. The Genes_seq track shows the exons implied by the composite alignment of transcripts, such as NCBI mRNA RefSeqs, to the genome. There are two boxed regions in the Map Viewer display. The upper boxed region shows that the GenomeScan model for FMR1 begins at a point that is part of an intron in the transcript-based model. Furthermore, there is a separate GenomScan model upstream of the first that overlaps with the initial exon of the transcript-based model. It would be of interest to investigate whether the two GenomeScan models could be fused to produce a longer transcript. In the lower boxed region, we see that the transcript-based model includes an exon lacking in the GenomeScan model. Perhaps we can create a model transcript based on the fusion of the two GenomeScan models that also includes the extra exon seen in the transcript-based model.
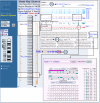
Figure 5
Use of ModelMaker to test alternative cDNA XSmodels based on GenomeScan predictions of mRNA alignments.
To attempt this synthesis, we first select the mm link [(a)] to the right of the FMR1 gene link on the Genes_seq track to invoke the ModelMaker. A number of alignments between transcript or model sequences and the genomic contig upon which FMR1 lies, NT_011537 [(b)], are given at the top of the ModelMaker display, and the implied exons resulting from these alignments are shown just below, numbered sequentially in the Putative exons pallet. We may choose any of these exons for inclusion in our model by selecting it. We can also choose a complete set of exons from an existing alignment by selecting the set link next to an alignment. Because we plan to begin with the GenomeScan model, we can select the set link next to the second alignment from the bottom to start. This gives us an initial model identical to that of the GenomeScan prediction and yields, as its longest Open Reading Frame (ORF), an ORF of 600 amino acids. We can also see the second, small, GenomeScan-predicted model at the far left of the ModelMaker display. We hope to fuse this model with the larger model. The second model comprises three closely spaced exons, resembling a single exon in the display, numbered 2–4 in the Putative exons pallet. Selecting exons 2 and 3 adds them to the model (first boxed pair in the ModelMaker display). At this point, the longest ORF detected in the transcript has increased to 762 amino acids. If we try to include exon 4, however, we drop back to 600 amino acids because of the introduction of an internal stop codon; therefore, we can remove exon 4 from our model with another click. Finally, we can select exon 22, which is the exon from the alignment-based model that we want to include, and notice that the longest ORF detected has risen to 823 amino acids [(c)]. Because long ORFs without stop codons occur rarely by chance, this transcript model is promising. To explore further, we can select the ORF Finder link to generate a graphical view of all ORFs found in the transcript, including the longest ORF, and subject the translation of the latter to a BLAST search. In this case, we find that the majority of the predicted protein matches the FMR1 gene product but that we have introduced some novel peptide sequence at the amino-terminal end as well as some near the carboxy terminus.
In this example, there were multiple, putative, full-length mRNAs. Please note that ESTs can be added to the display by selecting add ESTs (Figure 5, upper right-hand corner). Additional exons and splicing patterns may then be available to be considered in your model. This feature may be of particular importance if most of the evidence for splicing and exons comes from ESTs rather than complete mRNAs.
6. Using the Mouse Map Viewer
This assembly can be displayed by using Map Viewer for the mouse. In this particular case, the official symbol for the human and mouse genes is the same; therefore, a query by symbol returns the expected result. If this were not the case, however, it is also possible to search the mouse genome by the human sequence using BLAST the Mouse Genome. Simulating this, you can see that the best match for the human sequence is for a gene labeled LOC207836. If you check this in LocusLink, you will see that this is now annotated as Fmr1.
By Human/Mouse Homology Map
Consider an example in which we want to know whether there is a human/mouse synteny region that includes our favorite gene, FMR1. To begin, enter FMR1 into the Map Viewer search box and press the Find button. Selecting the gene name in the results table leads us to a Map Viewer display of the FMR1 gene with the Genes_seq map, UniG_Hs map, and Genes_cyto maps shown. In Figure 6, the Genes_cyto map has been replaced with the UniG_Mm map. Selecting the hm link (boxed) to the right of the Genes_seq map leads to the human/mouse homology maps. One can choose between two slightly different variants of human genome assembly (NCBI and UCSC). Three sources of mouse mapping data are available: the Mouse Genome Database (MGD) map, Jackson Lab map, and the Whitehead/MRC Radiation Hybrid map. Either of the two human maps may be compared with either of the two mouse maps.
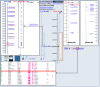
Figure 6
The human/mouse synteny region.
Let us choose the NCBI versus MGD variant and then check for synteny in the region of the FMR1 homologs in the two species. Because our primary interest is the human gene FMR1, we select NCBI versus MGD_Hs, and the comparative map will appear. The row corresponding to the FMR1 gene is highlighted (inset). The mouse homolog, called Fmr1, is also positioned on chromosome X, and the order of genes surrounding it is conserved in both genomes. Available STS data are indicated by a green dot that is a hyperlink to UniSTS. Links to the Map Viewer (select Cytogenetic map position) and LocusLink (select Gene Symbol) are available for each pair of genes. The user can examine the pairwise BLAST alignment that is accessible by selecting a chromosome color-coded dot preceding the mouse gene name. A dot-matrix similarity plot on the pairwise alignment page makes it easy to visually estimate the differences in sequences of the two genes. It is interesting to see that the similarity between FMR1 and Fmr1 actually extends past the coding regions.
Using the Mouse UniGene Map
Figure 6 shows a Map Viewer display of human FMR1 using the Genes_seq map, the UniG_Mm map, and the UniG_Hs map. It is apparent that the EST mapping patterns on the two UniGene maps are similar for FMR1. This indicates a similarity at the expressed sequence level but does not indicate a similar gene structure because both sets of ESTs are being mapped to the same human genomic sequence. To investigate similarities in gene structure, it is necessary to use the mouse version of the Map Viewer.
Using the Mouse Map Viewer
The most complete assembly of the mouse genome available at NCBI is the Mouse Genome Consortium Version 3 WGS assembly. This assembly can be displayed using the mouse version of the Map Viewer; however, the mouse homolog of FMR1 is not labeled as such in this assembly; therefore, we cannot find it using a simple search by gene name. We can overcome this obstacle by searching the mouse genome with the human mRNA sequence using mouse genome BLAST. The result of such a search is shown in the leftmost inset in Figure 6 and indicates that a gene model labeled LOC207836 is the probable homolog. The double-arrow links the human and mouse Map Viewer displays of the corresponding genes in the two species; the structures are not identical, but they do share a rather large 3′ exon. If we zoom out a bit in the two displays (two right insets) to see the surrounding genes, we can observe that the organization of the two genomes is similar in the region of FMR1 to the extent that a second gene called FMR2 lies downstream of FMR1, whereas the mouse version, Fmr2, likewise lies downstream of the mouse Fmr1 homolog.
7. How Can I Find Members of a Gene Family Using the Map Viewer?
Finding members of a gene family is not straightforward by any means. However, the Map Viewer can be used to flag sets of genes that are related, either by nomenclature or by sequence similarity.
By Common Annotation
Consider the gene FMR1. Let us assume we do not know much about genes from this family, but we suppose (recognizing that we may have cause to regret this supposition) that they all share the common root name FMR. We start our search from the main Map Viewer page by entering FMR* (the asterisk is a wild-card symbol) into the Search box and pressing the Find button. This search results in several hits, and some obviously do not belong to the FMR1 family (cytoplasmic FMR1 interacting proteins 1 and 2, FMRFAL); but two genes on chromosome X (FMR2 and FMR3) and one on chromosome 17 (FMR1L2) look promising. By selecting the chromosome X all matches link, we go to the graphical representation of the genomic region containing three FMR genes.
Selecting the gene name (the rows for the FMR* query hits are highlighted) invokes a corresponding LocusLink page that serves as a portal to available information for the gene, including the precomputed results of a similarity search against the nr database. Go to the NCBI Reference Sequences section of the LocusLink page and select the BLAST Link (BL) link. BLink displays a schematic representation of BLAST alignments with links to displays of the best hit from each organism, protein domains found in the query sequence, or sequences similar to the query that have known 3D structures.
When we look at the BLAST summary for FMR1, we find neither FMR2 nor FMR3. We did not expect FMR3 to show up because it had not been mapped on the Genes_seq map, which suggested that its sequence is not yet known. However, why do we not see FMR2? If we use LocusLink to retrieve the Reference Sequences for the FMR1 and FMR2 gene products (NP_002015 and NP_002016) and perform a pairwise BLAST comparison, we find that there is no significant similarity between the two sequences. Apparently, the names of “FMR” genes do not reflect common sequence features but rather a physiological condition, “fragile X mental retardation” syndrome, associated with this gene. In this sense, the two are members of a group or family, but they show sequence similarity only in the pathological trinucleotide repeats (CGG)n that are often found upstream of their coding regions.
By Precomputed Sequence Similarity
The BLink page lists two annotated human homologs of the FMR1 gene, called “fragile X mental retardation”, autosomal homologs 1 and 2 (FXR1 and FXR2). They show a significant similarity to the FMR1 protein and possess the same functional domains (K-homology RNA-binding domains documented in the LocusLink reports). Selecting the Score column link invokes a page with the results of pairwise BLAST, as well as a visual representation of similarity between the two proteins (a variant of the dot matrix similarity plot).
By Sequence Similarity to a Query
To see whether there are undocumented homologs of FMR1 in the genome, return to the Map Viewer maps page for the FMR1 gene and select the BLAST the Human Genome link. Enter the Accession number for the FMR1 protein taken from the LocusLink report (NP_002015) into the Search window, select Genome as the database, tblastn as the BLAST program, and search. A tblastn search takes a protein sequence as a query and translates a nucleotide database in all reading frames to find any coding regions, documented or undocumented, that might code for a protein similar to the query. Such a search is very sensitive because it is tolerant of differences in codon usage as well as of insertions and deletions.
The results of such a search are shown in Figure 7. BLAST hits to regions on four chromosomes are shown in the genomic overview, indicated by small tick marks. There are 14 hits to chromosome X, clustered near the end of the q-arm. These hits are to FMR1, which is located in band Xq27.3. There are also 5 hits apiece to chromosomes 3 and 17 [(a) and (b), respectively]. These hits are to known homologs of FMR1, FXR1, and FXR2. Selecting the links below the chromosome graphic or on the appropriate contig link in the table below leads to a graphical display of the hits on the contig, as shown in the two insets. Note that the BLAST hits track the exons of the genes. The hit on chromosome 12 is to a hypothetical protein and may be worth further investigation.
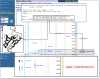
Figure 8
BLAST hits.
8. How Can I Find Genes Encoding a Protein Domain Using the Map Viewer?
The Map Viewer displays a graphic representation of genomic information with links to related resources that allow it to serve as a springboard for many types of analyses.
Finding Protein Domains in a Gene Product
For example, one might be interested in the domain structure of the gene product. Let us use the human FMR1 gene as an example. On the main Map Viewer page, enter FMR1 into the Search for window and press the Find button. Select the FMR1 link to display three maps (cyto, UniG_Hs, and Genes_seq) of the FMR1 locus on chromosome X. Selecting the gene name next to the gene model (FMR1) leads to the LocusLink page, which is not only a compilation of genomic, genetic, and reference data related to the gene but also a gateway to the external resources and NCBI tools and databases. The LocusLink report for FMR1 is linked to precomputed BLAST results for the FMR1 gene product. Selecting BL (BLink, for BLAST link) invokes a page with a schematic view of the BLAST comparison of the FMR1 protein against the non-redundant (nr) database. The BLink page lists database hits to the FMR1 protein, sorted according to their BLAST similarity scores. Results may be reformatted by selecting Sort by Taxonomy Proximity to cluster hits from the same species.
To see the functional domains that have been identified in the FMR1 protein, select the Conserved Domains Database (CDD) Search button. The resulting page shows two hits to the KH-domain from the SMART database and one hit to the KH domain in the Pfam database. Notice that one of the SMART domain hits is a partial hit, as indicated by the jagged edge in the schematic representation.
Visualizing 3D structures
We can easily see whether there exists a three-dimensional structure that includes these conserved domains. The pink dot preceding the domain name in the BLink Description of Alignments section leads to a display of the corresponding three-dimensional structure using Cn3D, the NCBI macromolecular viewer available over the Web. The Pfam and SMART domains are linked to two 3D structures (1K1G_A and 1J4W_A). The CDD page also lists other sequences that have the same domain and shows their multiple sequence alignments.
Searching for Similar Domains in Genomic Sequence
Cut and paste the sequence of the KH-domain from the FMR1 protein and run a genome-specific BLAST search. We will use the tblastn program to compare the 44-amino acid sequence of the KH domain to the nucleotide sequence of the human genome. The results will show us other regions of the genome with the potential to code for this domain. Of course, we already know from the BLink page that there are some autosomal homologs, but we do not know whether they actually contain the KH RNA-binding domain. The obvious caveat is that some pseudogenes might contain the domain as well. Our tblastn search returns 4 hits, one being the FMR1 gene itself on the X chromosome, and three others on chromosomes 3, 12, and 17. Select the Genome View button in the Genome BLAST results to see the positions of the hits on the chromosomes. One may then select the names of sequences producing significant alignments to invoke a Map Viewer display that shows the corresponding maps and report the names of the loci. As expected, hits to chromosomes 3 and 17 correspond to the autosomal homologs FXR1 and FXR2. The hit to chromosome 12 corresponds to the hypothetical protein HSPC232, and the Map Viewer display for this BLAST hit is shown (Figure 8). The hit is to a segment of intronic sequence, rather than to an exon, and is without supporting human EST alignments. There are, however, mouse ESTs mapping to this region (lower inset), and there is also a GenomeScan model that covers the hit; therefore, the BLAST hit may indeed represent coding sequence. Selecting the Blast hit link (boxed) leads to an alignment (upper inset) that indicates a good match between our 44-amino acid domain sequence and a protein translation of the genomic sequence. If we map this 44-amino acid sequence onto the structure of 1K1G using Cn3D, we see that it covers a module consisting of two alpha helices and a three-stranded beta sheet (leftmost inset). It appears reasonable that our domain hit may represent an exon of the HSPC232 gene. To follow this line of analysis, the next step might be to produce a transcript model incorporating this new exon using the Model Maker (see the Model Maker exercise in this series).
- Introduction
- 1. How Do I Obtain the Genomic Sequence around My Gene of Interest?
- 2. If I Have Physical and/or Genetic Mapping Data, How Do I Use the Map Viewer to Find a Candidate Disease Gene in That Region?
- 3. How Can I Find and Display a Gene with the Map Viewer?
- 4. How Can I Analyze a Gene Using the Map Viewer?
- 5. How Can I Create My Own Transcript Models with the Map Viewer?
- 6. Using the Mouse Map Viewer
- 7. How Can I Find Members of a Gene Family Using the Map Viewer?
- 8. How Can I Find Genes Encoding a Protein Domain Using the Map Viewer?
- Exercises: Using Map Viewer - The NCBI HandbookExercises: Using Map Viewer - The NCBI Handbook
Your browsing activity is empty.
Activity recording is turned off.
See more...