NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Featured Resource: The NCBI Short Read Archive (SRA) of Next- Generation Sequencing Data
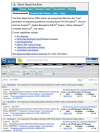
The NCBI now maintains the Short Read Archive (SRA) (www.ncbi.nlm.nih.gov/Traces/sra/) as a repository for data from sequencing projects that use the new massively parallel sequencing technologies, often called next-generation sequencing. These methods can generate hundreds of megabases to gigabases of data in a single instrument run, millions of times the output of a standard Sanger sequencing instrument. Applications of these technologies include sequencing of new genomes, re-sequencing of targeted genomic regions, sequencing complete genomes of multiple individuals to mine for variations, transcriptome sequencing to sample splice variants and expression levels, environmental samples and other metagenome sequencing, and chromatin DNA binding protein analysis. SRA provides the ability to search and display aspects of SRA project data through the SRA homepage (Figure 1, top panel), and the Entrez system (Figure 1, bottom panel. The SRA site also provides direct access to download data through the Aspera Connect (www.aspera.com) client that offers much faster transfers than traditional ftp. A recently added BLAST service allows searches against the transcriptome sequencing studies from the SRA data.
The Short Read Archive will become quite important as next-generation sequencing technologies continue to improve and become even less expensive. The power and capabilities of the SRA site will expand to provide better and more powerful options for searching and connecting these data to other resources.
Next-Generation Sequencing Technologies
SRA accepts and presents data from all current next-generation sequencing platforms including 454 (Roche), Illumina, SOLiD (Applied Biosystems), HeliScope, and Complete Genomics. While these systems use different approaches to isolate and amplify the target molecules and to generate sequence, all rely on extreme miniaturization of the system components, simultaneous reactions in parallel in a flow cell, light-based detection of in the sequencing reactions, and image analysis to acquire sequence information from multiple reactions at once. These methods yield huge numbers of short sequence reads from a single instrument run. Individual read lengths vary from around 25 bases to more than 400 bases depending on the platform. Data can include sequence, quality scores, color values, and intensity graphs depending on the platform involved.
Data in SRA
Data Concepts
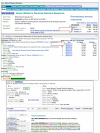
Data in the SRA are classified into a hierarchy of Studies, Experiments, Samples, and their corresponding Runs. Studies have an overall goal and may be comprised of several Experiments. An Experiment describes specifically what was sequenced and the method used. It includes information about the source of the DNA, the Sample, the sequencing platform, and the processing of the data. Each Experiment is made up of one or more instrument Runs. A Run contains the results or reads from each spot in the instrument run. In the future, some data will also have an associated Analysis. These Analyses may include assemblies of the short reads into genomic or transcript contigs and alignment to existing genomes or alignments with SRA data. Records at each level have unique accession identifiers with a specific three letter prefix that indicates the type of record: ERP or SRP for Studies, SRS for samples, SRX for Experiments, and SRR for Runs. Figure 2 shows Study (SRP000095, top panel), Experiment (SRX000113, middle panel, and SRX000114), and Run (SRR000416, bottom panel) records for the 454 sequencing of James Watson’s genome by Cold Spring Harbor Laboratory. Study and Run records are displayed in the SRA browser. The corresponding Experiment records are displayed in the NCBI Entrez system as described in the next section.
Searching and Viewing SRA Data in the SRA Browser and Entrez
Studies, Runs, and their associated Samples can be viewed and browsed through the SRA browser link on the SRA homepage.
www.ncbi.nlm.nih.gov/Traces/sra
Experiment records are available for searching in the Entrez SRA database.
www.ncbi.nlm.nih.gov/sites/entrez?db=sra
As with other Entrez databases using field limits in search queries produce more precise results. The organism field is useful, as in all NCBI molecular databases, for finding experiments involving a particular taxon. The properties field is helpful for finding specific types of SRA studies. For example, the following query finds all human genomic resequencing Experiments – 984 at the time of this writing.
human[organism] AND study type resequencing[Properties] AND biomol genomic[Properties]
All of the available fields and their indexed terms can be browsed through the Preview/Index tab on the SRA Entrez search page.
The record for the Study associated with an Experiment, all Experiments for the Study, and Experiments that used the same sample are easily retrieved through links on the Entrez SRA Experiment record (Figure 2, middle panel). SRA Experiment records in Entrez are integrated with data from other Entrez databases. Links to PubMed, GEO datasets, Genome Projects, Nucleotide, and Taxonomy are currently available for the Experiment records. Currently 6,240 Experiments are available from 806 Studies.
SRA BLAST Service
In addition to text searches of the SRA experiments through Entrez, NCBI also offers a nucleotide BLAST service for sequence similarity searching of 454 sequencing reads for transcriptome studies. This service is accessible from the “Specialized BLAST” section of the BLAST Homepage.
Databases are labeled by taxon. Currently there are transcriptome reads for 31 species and two metagenome data sets.
Downloading SRA Data
SRA data can be downloaded through the “Download” tab on the SRA homepage or through the Download link that is present on Study, Sample, and Experiment records (Figure 2). Because data for SRA projects often exceed 10 Gigabytes, traditional ftp may be too slow to download data effectively. To avoid this problem, SRA download links use the fasptm protocol developed by Aspera to transfer data. This protocol is more efficient and stable than traditional ftp. The free Aspera Connect Web browser plug-in, available from the company’s Website, is required to download SRA data.
Once installed Aspera Connect will launch to transfer data from SRA whenever a download link is clicked. SRA offers standard FASTA and the convenient and portable fastq format for download. The fastq format is ASCII text that includes the sequence plus the ASCII encoded quality scores.
Submitting Data to SRA
SRA provides an interactive web-based interface for submissions that requires only a brief registration prior to submission. The Submissions tab on the SRA homepage accesses the registration and login page for SRA submissions (Figure 1, top panel). SRA also offers an automated submission pipeline for centers making multiple submissions. Detailed information on submitting to SRA is available in the SRA Submission Guidelines document.
www.ncbi.nlm.nih.gov/Traces/sra/static/SRA_Submission_Guidelines.pdf
Summary
SRA data are rapidly dominating all other sequence data. Already the number of DNA bases available in SRA exceeds the number of bases in GenBank. In fact the output of a single important project, the 1000 genomes project (www.1000genomes.org), will produce more than 25 times the number of bases that are currently in GenBank by the time the project is completed. The NCBI and SRA will continue to support submission, retrieval, and analyses of these increasingly challenging and complex sequencing data. Means of displaying data, analyses, and integration of SRA data with other molecular databases will continue to improve making the SRA data a prominent part of the discovery system at the NCBI.
New Databases and Tools
Human Genome Build 37.1
Human genome build 37.1, the new Human Genome Reference Consortium assembly and annotation, is now displayed in the NCBI Entrez system and the NCBI Map Viewer site.
GenBank News
GenBank release 172.0 is incorporated into the NCBI and FTP sites (ftp.ncbi.nih.gov/genbank/). The current release includes data available as of June 10, 2009. Release notes (gbrel.txt) describing details of the release and upcoming changes are in the GenBank FTP directory.
NCBI is considering discontinuing the index files; affected users are encouraged to review the discussion of this change in the release notes and provide comments to the GenBank group.
Updates and Enhancements
HomoloGene
HomoloGene release 64 includes updated annotations for Homo sapiens (NCBI release 37.1), Caenorhabditis elegans (WS190, NCBI release 8.1), Anopheles gambiae (AgamP3.3, NCBI release 3.1), Arabidopsis thaliana (NCBI release 8.1), Bos taurus (NCBI release 3.1), and Magnaporthe grisea (NCBI release 3.1). The HomoloGene homepage has additional details.
RefSeq
RefSeq Release 36, now available through NCBI Entrez and FTP (ftp.ncbi.nlm.nih.gov/refseq/release/) incorporates genomic, transcript, and protein data available as of July 2, 2009. It includes 12,141,825 records from 8,665 different species and strains. Changes since the previous release are described in the notes in the RefSeq FTP directory.
BLAST
With the new BLAST 2.2.21 release, the BLAST+ command-line applications, written with the NCBI C++ toolbox, are now the major supported version of BLAST. The BLAST+ applications have a number of advantages over the older applications that include working more robustly with long sequences and database masking. The BLAST+ applications were described in the January 2009 NCBI News. The FTP directory contains a complete user manual for the BLAST+ package.
ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/user_manual.pdf
Influenza Virus Resource
The Influenza Virus Resource has an option for viewing “Sequences from Pandemic (H1N1) 2009 virus only” on the database search page.
www.ncbi.nlm.nih.gov/genomes/FLU/Database/select.cgi?go=1
The page also offers an option to exclude these sequences from search results if desired.
PubMed Central
Are you interested in new titles added to PubMed Central? If so, the PMC RSS feed provides all new article titles as well as titles of newly scanned articles from archives.
Announce Lists and RSS Feeds
Fifteen topic-specific mailing lists are available which provide email announcements about changes and updates to NCBI resources including dbGaP, BLAST, GenBank, and Sequin. The various lists are described on the Announcement List summary page: www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html. To receive updates on the NCBI News, please see: www.ncbi.nlm.nih.gov/About/news/announce_submit.html
Seven RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, PubChem, LinkOut, HomoloGene, and NCBI Announce. Please see: www.ncbi.nlm.nih.gov/feed/
Comments and questions about NCBI resources may be sent to NCBI at: vog.hin.mln.ibcn@ofni, or by calling 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
- NCBI News, August 2009 - NCBI NewsNCBI News, August 2009 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...