NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Featured Resource: The BioSystems Database of Biological Pathways
NCBI BioSystems is a new database that collects information on interacting sets of biomolecules involved in metabolic and signaling pathways, disease states, and other biological processes. BioSystems currently contains biological pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) and the EcoCyc (Escherichia coli K-12 MG1655) subset of the BioCyc databases and is designed to accommodate other data in the future. BioSystems is fully integrated with other databases in the Entrez system with links to related literature, genes, protein sequences, structures, chemical data, and to related BioSystems. Along with links to related data at the NCBI site, each BioSystem record provides links to detailed diagrams and annotations for individual pathways on the Web sites of the source databases. BioSystems adds an important new aspect to the NCBI system by linking many different kinds of molecular records in biochemical pathways and providing means to compare these pathways across organisms.
Searching BioSystems
BioSystems is available as part of the NCBI Entrez system and can be searched directly from the database page:
www.ncbi.nlm.nih.gov/BioSystems/
As with other Entrez databases, field-restricted queries in BioSystems give more precise search results. Many of the fields that work in the other Entrez molecular databases such as Organism and Title also are useful in BioSystems. All of the available fields can be browsed through the Preview/Index tab on the search page. The Limits tab provides a simple way to limit to certain types of records through pre-set fields. For example, the search can be limited to a specific source database, currently KEGG or EcoCyc. There is also a checkbox that allows restricting to organism-specific BioSystems, those with links to molecular records for a single species, or conserved BioSystems that group together orthologous organism specific pathways derived from KEGG reference pathways. Limiting to Conserved BioSystems provides more concise results and access to an overall summary of the pathway in all organisms. For example, a search with the phrase “purine metabolism” retrieves 3,509 records while limiting to conserved BioSystems or clicking the Conserved BioSystems filter tab provides a more concise set of only 26 records (Figure 1). As another example, the organism-specific record for purine metabolism in mouse is found directly with the following field-restricted query.
purine metabolism[Title] AND mouse[Organism]
BioSystems Records: Photosynthesis
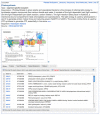
The BioSystems database currently contains over 95 thousand pathways from the KEGG database including 255 conserved BioSystems and 286 Escherichia coli K-12 pathways from the EcoCyc portion of the BioCyc database. Figure 2 shows the BioSystems organism-specific record for photosynthesis from Arabidopsis thaliana (BioSystems ID 4001). The record has a description of the process from the source database, a thumbnail graphic that links to the larger diagram in the KEGG database, and a Tabbed Table of components and other aspects of the pathway (Genes, Proteins, Small Molecules, Related BioSystems, Citations and Comments). These components link to records in the NCBI Gene, Protein, PubChem, BioSystems, and PubMed databases.
Pathway Diagram
The full photosynthesis diagram is available at the KEGG site and is linked to the thumbnail image in the BioSystems record. The cartoon of the chloroplast thylakoid membrane at the KEGG site shows the multi-subunit protein complexes of the photosynthetic system (photosystems I and II, cytochrome b6/f, electron transport system, and the f-type ATPase). The boxes in the tables below the cartoon represent the genes or protein components of the system. Each of the green-filled boxes represents one or more Arabidopsis genes. The unfilled boxes represent genes or functions that are not known from Arabidopsis. In this case these missing components (PsbU, PsbV, PsbX, Psb28-2, PsaM, PsaX, PetL, and PetM) are specific to certain cyanobacterial systems and are available in the corresponding Conserved BioSystem or reference pathway.
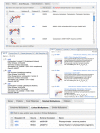
Tabbed Table of BioSystem Components
The Tabbed Table provides access to the components and other information linked to the pathway in the NCBI databases. Tabs include Genes, Proteins, Small Molecules, Related BioSystems, and Citations. The Arabidopsis genes and proteins corresponding to the filled boxes in the pathway diagram are listed in the Genes and Proteins tabbed sections of the table. The Small Molecule tab provides access to substrates, inhibitors, cofactors, and other non-protein entities involved in pathways from the NCBI PubChem database. Clicking on any entry will link to the corresponding Gene, Protein, or PubChem record. The link at the top of the table retrieves all records in the active tab. For example, clicking the top link in the Genes tab easily retrieves all 73 Arabidopsis genes involved in the photosynthetic pathway; clicking this link in the Small Molecules tab retrieves cofactors and substrates from PubChem (Figure 3). The Related BioSystems tab expands to display three types of related BioSystems: Linked BioSystems, Similar BioSystems, and Conserved BioSystems. Linked BioSystems are pathways that interact with the current pathway often providing substrates or receiving products of the current pathway. In this case, these are the antennal proteins light collecting system of photosynthesis (BioSystems ID 4002) and carbon fixation in photosynthetic organisms (BioSystems ID 4065). Similar BioSystems are pathways that share at least one identical protein sequence from the same source organism. Oxidative phosphorylation (BioSystems ID 4000) is the Similar Pathway to photosynthesis because it shares the subunits of the f-type ATPase. Finally, the Conserved BioSystem (BioSystems ID 340) is the reference pathway record for photosynthesis that gathers all orthologous pathways. The Conserved BioSystem includes the cyanobacterial components represented by unfilled boxes in the organism-specific pathway at the KEGG site. The remaining populated tab for the photosynthesis record, Citations, provides literature citations from the source database with links to the corresponding records in PubMed.
Links Menus: Related Data
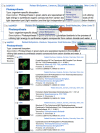
In addition to the linked data available in the Tabbed Table, each BioSystems record has related data available as Links menus at the upper right of the summary or full BioSystems record (Figure 4). These are the Related BioSystems, Literature, Sequences, Small Molecules, and Other Links menus. The BioSystems, Literature, and Small Molecules menus access the same related data available in the Tabbed Table. Additional related data in the Sequences menu that are not available in the Tabbed Table are HomoloGene, Protein Clusters, and Conserved Domain records that are linked from the proteins in the pathway.
The HomoloGene and Protein Clusters related data identify homologs from selected eukaryotic and microbial genomes and allow comparisons of pathways across taxa. The linked Conserved Domains can give additional structural and functional information about the proteins in the pathway. The Other Links menu connects the BioSystem to the PubChem BioAssay database for small molecules in the pathway that have been classified as active in one of the assays. There are also links to the NCBI taxonomy database for organism-specific BioSystems, and to the Structure database if any of the proteins in the pathway are linked to a three-dimensional structure. The following example shows how to use these related data to find structures for the photosynthetic complexes starting from the Arabipdopsis organism-specific record.
Example: Using Related Data to Find Three-Dimensional Structures
The links to data available through the Tabbed Table and Links menus can be used to find important related data such as homologous pathways, genes, and proteins in other species as well additional structural and functional information. For example, using the Tabbed Table and the Links menus, it is easy to find the three-dimensional structures of some of the protein complexes involved in photosynthesis. As mentioned previously, structure records are linked to the BioSystems pathway through the “Other Links” menu. However, the Arabidopsis photosynthesis pathway has no direct structure links because none of the photosynthesis proteins with structures are from this species. Following the Related BioSystems link to the Conserved BioSystem gives access to proteins from orthologous pathways in all species including those with structure records. The Conserved BioSystem for photosynthesis (BioSystems ID 340) has links to five structures of photosynthetic protein complexes through the Other Links menu (Figure 4). Four of these structures are from cyanobacteria, and one is from the green alga, Chlamydomonas reinhardtii.
Summary
The BioSystems database adds a new dimension to NCBI resources by connecting molecular, bioassay, and literature data to biological pathways and processes. This enables searches that for the first time can find all gene, protein records, and small molecules in a pathway for a particular organism. Moreover, integration with other NCBI databases allows comparisons of pathways across taxa and provides access to other data on structure and function of the biomolecules involved. Expanding the coverage of the Entrez system to pathways provides new connections among databases that should greatly increase the power of Entrez as a discovery system.
New Databases and Tools
New BankIt Submission Tool
A new version of the BankIt sequence submission tool is available for testing. The new tool will eventually replace current version of BankIt after the test period. Please see the New BankIt page for more information: www.ncbi.nlm.nih.gov/WebSub/?tool=genbank
Bookshelf
The book, Familial Cancer Syndromes has been added to the NCBI Bookshelf. To browse this book go to www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=famcan
Microbial Genomes
Twenty-one finished microbial genomes were released between May 29 and July 6, 2009. The original sequence data files submitted to GenBank/EMBL/DDBJ are available on the FTP site: ftp.ncbi.nih.gov/genbank/genomes/Bacteria/. The RefSeq provisional versions of these genomes are also available: ftp.ncbi.nih.gov/genomes/Bacteria/.
GenBank News
GenBank release 172.0 is available on the NCBI Web and FTP sites. The current release includes information available as of June 10, 2009. Release notes are available on the on the NCBI ftp site: ftp.ncbi.nih.gov/genbank/gbrel.txt
NCBI is considering ceasing support for the index files; affected users are encouraged to review the discussion of this change in the release notes and provide comments to the GenBank group.
Updates and Enhancements
PubChem
The American Library Association has selected PubChem as one of the Best Free Reference Web Sites 2009. More PubChem announcements are available at: pubchem.ncbi.nlm.nih.gov/pcnews.html and as an RSS feed.
BLAST
As previewed in the May 2009 NCBI News, COBALT multiple-sequence alignments can now be generated from protein BLAST results by clicking on the “Multiple Alignment” link. A direct submission form for generating protein multiple alignments using COBALT is also available in the Specialized BLAST section of the BLAST Homepage. The BLAST News page provides additional details about COBALT. The BLAST homepage also provides a “Tip of the Day” for more efficient use of the BLAST tool.
Announce Lists and RSS Feeds
Fifteen topic-specific mailing lists are available which provide email announcements about changes and updates to NCBI resources including dbGaP, BLAST, GenBank, and Sequin. The various lists are described on the Announcement List summary page: www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html. To receive updates on the NCBI News, please see: www.ncbi.nlm.nih.gov/About/news/announce_submit.html
Seven RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, PubChem, LinkOut, HomoloGene, and NCBI Announce. Please see: www.ncbi.nlm.nih.gov/feed/
Comments and questions about NCBI resources may be sent to NCBI at: vog.hin.mln.ibcn@ofni, or by calling 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
- NCBI News, July 2009 - NCBI NewsNCBI News, July 2009 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...