NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Featured Resource: The Genome Reference Consortium Human Genome Build 37 now Available
In August the NCBI released the annotation of build 37 of the human genome. This build includes new sequence and assembly provided by the Genome Reference Consortium (GRC). The GRC is a collaboration of the Wellcome Trust Sanger Center, the Washington University Genome Center, the European Bioinformatics Institute and the NCBI. The goal of the GRC is to correct misassembled regions, to close remaining gaps, and to provide alternate assemblies of structurally variant positions (loci) in the genome. Build 37, also known as GRCh37, includes updates for all human chromosomes, closes 25 sequence gaps, corrects over 150 problems in build 36, and adds nine alternate loci.
The GRC page at NCBI provides additional details about this new assembly.
www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human/
The NCBI Website provides easy access for searching and exploring the sequences and annotations of the new and improved primary reference genome and alternate loci through the Entrez system, the graphical sequence viewer, the Map Viewer, and the NCBI Web BLAST services.
GRCh37 Sequences at NCBI
The GRCh37 assembly includes the assembled human chromosomes, some unlocalized and unplaced sequence, and alternate assemblies for structurally variable regions in the genome. The primary assembly chromosome sequences are available under accession numbers CM000663 through CM000686. These are assemblies of the 22 autosomes plus the X and Y chromosomes. The nine alternate assemblies are for the following regions: the UDP glucuronosyltransferase 2, polypeptide B17 gene (UGT2B17) on chromosome 4 (accession GL000257); the Major Histocompatibility Complex (MHC) on chromosome 6 (accessions GL000250 through GL000256); and the microtubule-associated protein tau (MAPT) gene on chromosome 17 (accession GL000258).
The NCBI genome annotation pipeline has created a corresponding set of 31 reference sequences (RefSeqs) that provide the locations of genes and other features on the GRCh37 reference assembly and alternate loci. Table 1 shows the correspondence between the RefSeq and GenBank records for GRCh37.
Table 1
Correspondence of GenBank, RefSeq accession numbers, and assembled sequences for the GRCh37 reference genome.
Retrieving and Viewing CRCh37 at NCBI
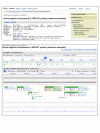
GRCh37 sequences and annotations are easily retrieved and viewed in the Entrez system and the NCBI Map Viewer. A search for GRCh37[Title] in the Entrez nucleotide database (www.ncbi.nlm.nih.gov/sites/entrez?db=nuccore) collects all 564 records associated with the current build. Restricting to reference sequences (ReSeq) using the filter tab limits the results to the 282 processed RefSeq versions of chromosomes, contigs, and alternate loci that include the annotations of biological features. Figure 1 (top panel) shows the traditional GenBank view of the GRCh37 chromosome 4 (NC_000004) in the Entrez system. This abbreviated view can be adjusted with controls on the page to add biological features and sequence. However, the large number of features and long sequence make this an awkward way to browse the data. The graphical sequence viewer, offered as the “Graphics report” link at the top of the GenBank view, provides a better alternative for exploring the chromosome record and its features. Following the Graphics report link and searching for the UGT2B17 as a marker results in the display of the region surrounding the UGT2B17 gene on chromosome 4 as shown in the bottom panel of Figure 1. The graphical viewer provides details of gene position structure and orientation, alignments of transcripts and proteins, and the ability to display SNPs and other markers. Each annotated gene or transcript in the graphical view has links to sequence display formats, other databases such as Gene, and the ability to run a BLAST search with the annotated genomic, transcript, or protein sequence (Figure 2).
The NCBI Map Viewer is another useful way to view aspects of the genome build. The human genome map viewer is accessible from the Map Viewer Homepage:
All genes, transcripts, and proteins associated with the genome have links to the build in the Map Viewer from the corresponding records in the Entrez system. In addition, the NCBI Web BLAST service can link results of searches against human genome plus transcript database as well as those from the separate human genome BLAST service directly into the Map Viewer. This BLAST search option can be used to highlight improvements in the human genome build as shown in the following example.
Example: Exploring Changes in Chromosome 4 in Build 37
As mentioned previously the GRCh37 assembly closed 25 gaps in the previous build (build 36) of the human genome. One such gap is in the region surrounding the UGT2B17 gene on chromosome 4. In build 36, this region appears to contain a partial duplication surrounding a gap. Since the human genome BLAST service and the Map Viewer allow searches against both GRCh37 and build 36, changes in the structure of this region between the two builds are easily demonstrated. Using the genomic region corresponding to the transmembrane serine protease 11E (TMPRSS11E) as a query in human genome BLAST (NC_000004, bases 69313167-69363322) shows the apparent duplication in build 36. This search is set-up directly from the TMPRSS11E gene in the graphical viewer by following the genome specific BLAST link from the Views and Tools pop-up menu (Figure 2, top panel, left). The results against build 36 show two near-perfect matches for the TMPRSS11E genomic region on different contigs flanking an apparent gap (Figure 2, top panel, right). This highlights an apparent duplication – but an incomplete one since the upper contig contains the UGT2B17 gene while the lower contig appears to lack this gene. This structure (duplication and gap) is known to be an artifact caused by the incorporation of two different alleles, one of which is a null allele for UGT2B17, into the build 36 genome (1). The current build solves this problem by incorporating the UGT2B17 containing allele into the primary reference genome and providing a separate record, ALT_REF_LOCI_8 (NT_167250), for the null allele. The structure of the new reference assembly and the alternate allele are easily demonstrated in the same way as for build 36 by a human genome BLAST search against build 37 (Figure 2, bottom panel).
Summary
The genome reference consortium (GRC) build 37 provides a more accurate and improved representation of the human genome by correcting errors, closing gaps, and providing alternate representations of structurally variant regions. The GRC itself, a collaboration among sequencing centers and bioinformatics resource and analysis centers such as the NCBI, will continue to provide the most up to date and accurate sequence and annotation for the reference human genome as additional data and analysis alter the view of the genome. The NCBI Website will continue to offer improved and more powerful visualization and analysis tools for investigating the human genome.
Reference
- 1.
- Xue Y Sun. Adaptive evolution of UGT2B17 copy-number. Adaptive evolution of UGT2B17 copy-number. 2008. pp. 337–46. [PMC free article: PMC2556428] [PubMed: 18760392]
New Databases and Tools
New NCBI Homepage
A new NCBI Homepage is available for beta testing during the next two months. The new look is cleaner and better organized than the current page. New features include a “How To” section for answers to common questions and links to resource lists. The new page is available for testing at the following URL:
http://preview.ncbi.nlm.nih.gov/guide/
Feedback is appreciated and encouraged. Please send feedback to vog.hin.mln.ibcn@ofni
PubMed Redesign
PubMed has also undergone reconstruction and is available for testing for a two week period. Many changes have been made that make search and retrieval easier and more comprehensive. The new design is quite different than the old but incorporates all of the new features that have been added over the past year such as Recent Activity, Ads, and Sensors. Please test the site and provide feedback on your experience.
http://preview.ncbi.nlm.nih.gov/pubmed
The National Library of Medicine Technical Bulletin provides a guide for making the transition to the new PubMed interface:
Rapid Research Notes
Rapid Research Notes (RRN) is a new resource that contains articles published online for immediate communication. The H1N1 outbreak prompted the development of RRN, but future collections will consist of other biomedical information as well. See the RRN homepage (www.ncbi.nlm.nih.gov/rrn/) and the “About” page (www.ncbi.nlm.nih.gov/rrn/about/index.html) for more information.
Microbial Genomes
Sixty-four finished microbial genomes were released during the dates July 1 - September 14. The original sequence data files submitted to GenBank/EMBL/DDBJ are available on the FTP site: ftp.ncbi.nih.gov/genbank/genomes/Bacteria/. The RefSeq provisional versions of these genomes are also available: ftp.ncbi.nih.gov/genomes/Bacteria/.
GenBank News
GenBank release 173.0 is now on the NCBI Web and FTP sites. The current release includes data available as of August 21, 2009. The release notes provide detailed information and statistics on the release: ftp.ncbi.nih.gov/genbank/gbrel.txt
Updates and Enhancements
RefSeq
RefSeq Release 37 is now part of the NCBI Entrez system and can be downloaded from the FTP site (ftp.ncbi.nlm.nih.gov/refseq/release/). This full release incorporates genomic, transcript, and protein data available as of September 3, 2009. It includes 12,941,750 records from 9,005 different species and strains. Changes since the previous release can be found in the release notes (ftp.ncbi.nlm.nih.gov/refseq/release/release-notes/RefSeq-release37.txt). More information on the RefSeq project is available on the RefSeq Homepage: www.ncbi.nlm.nih.gov/RefSeq/.
dbSNP
Complete data for the dbSNP Bovine build 130 are now part of the NCBI Entrez system and can be downloaded from the dbSNP FTP site. More detailed genome build information is available on the dbSNP page: www.ncbi.nlm.nih.gov/SNP/snp_summary.cgi.
Exhibits
NCBI will have an exhibit booth at the American Society of Human Genetics annual meeting in Honolulu, Hawaii, held October 20-24, 2009. Staff will present a tutorial, “The NCBI Discovery System: Integrated Access to Literature, Sequences, Genomes and Molecular Structures” on Wednesday, October 21 at 11:30 a.m. in the Convention Center (room 307).
Announce Lists and RSS Feeds
Three new mailing lists are available for updates and changes to NCBI resources. The new announce lists are: NCBI Structures, Conserved Domains, and BioSystems.
Eighteen topic-specific mailing lists are available which provide email announcements about changes and updates to NCBI resources including dbGaP, BLAST, GenBank, and Sequin. The various lists are described on the Announcement List summary page: www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html. For instructions on how to receive updates on the NCBI News, please visit: www.ncbi.nlm.nih.gov/About/news/announce_submit.html
Seven RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce. Please see: www.ncbi.nlm.nih.gov/feed/
Comments and questions about NCBI resources may be sent to NCBI at: vog.hin.mln.ibcn@ofni, or by calling 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
- NCBI News, September 2009 - NCBI NewsNCBI News, September 2009 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...