NCBI Discovery Workshops: Feb 15-16, 2011
NCBI will present a two-day workshop on February 15-16, on the NIH campus in Bethesda, Maryland. The course is free and is open to anyone interested in NCBI resources. The workshops provide hands-on experience exploring practical examples using tools and databases on the NCBI website. The four workshops are Sequences, Genomes, and Maps; Proteins, Domains and Structures; NCBI BLAST Services; and Human Variation and Disease Genes. For more information see the Discovery Workshop page, which also includes a registration link.
Updated Resources for Genomic Libraries and Clones
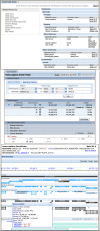
NCBI has updated resources for finding genomic libraries and genomic clones from genome sequencing projects for a large number or organisms. The new CloneDB (Figure 1, Top panel) replaces the Clone Registry as the resource for finding descriptions, sources, and detailed statistics on available genomic libraries for a large number of organisms.
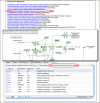
The new Library Browser (Figure 1, Bottom panel) allows filtering by organism, vector type, distributors, and number of associated database end or insert sequences. The linked Clone Finder (Figure 2), now available for human, mouse, rat, cow, horse, pig, and zebra finch, quickly identifies clones that span regions on assembled genomes.
The Clone Finder locates clones by chromosomal position or by features such as genes, SNPs, markers, or transcript sequence accession number. Clones may also be found in regions bounded by any two markers (Figure 2, Middle panel). The initial query may be refined to specific mapping data sets, population sources, and libraries. The graphical display in Clone Finder shows features annotated on the genome including assembled contigs, their components, genes, and aligned transcripts (Figure 2, Bottom panel).
Together CloneDB's Library browser and the Clone Finder provide essential access to these important molecular reagents.
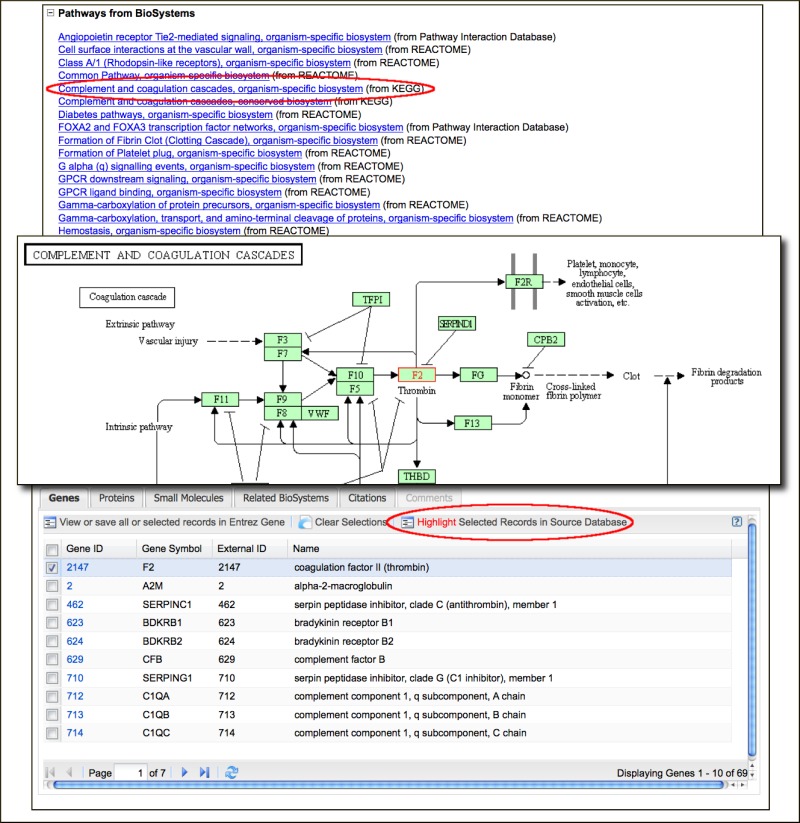
New Gene-BioSystems Links Highlight the Gene in the Biological Pathway
Links from BioSystems are now fully listed in a separate section of Gene records (Figure 3, Top panel). Each of these new links point to a specific pathway or system and leads to the record in BioSystems with the specific gene highlighted (Figure 3, Bottom panel). For pathways imported from the Kyoto Encyclopedia of Genes and Genomes (KEGG) the BioSystems record provides the option to highlight the record in the large pathway diagram at the KEGG site (Figure 3, Middle panel). A feature article in the July 2009 NCBI News describes the BioSystems resource in detail.
New Organisms in UniGene
Five new organisms have builds in UniGene: the hydrozoan (Cnidaria) Clytia haemisphaerica; the perigord truffle, Tuber melanosporum; the English or Truffle Oak, Quercus robur; the two-spotted spider mite, Tetranychus urticae; and the salmon louse, Lepeophtheirus salmonis.
Clytia haemisphaerica (Che build information, 4,637 clusters, FTP) is a marine hydrozoan in the phylum Cnidaria, the phylum that also contains jellyfish (scyphozoa) and corals (anthozoa). Unlike Hydra, the other hydrozoan in UniGene, Clytia has a free-swimming medusa stage. Gene and genomic information from Clytia has the potential to provide important insights on the evolution of animal body plans.
The perigord truffle (Tme build information, 7,543 clusters, FTP), an ascomycete fungus and the source of the gastronomically highly prized black truffle, and the truffle oak (Qro build information, 7,170 clusters, FTP) are two organisms linked in a symbiotic mycorrhizal association. UniGene sets from these two organisms should support sudies of genes involved in the evolution, function, and maintenance of symbiosis.
Two parasitic arthropods of economic importance also join UniGene. The two-spotted spider mite (Tur build information, 7,177 clusters, FTP) is a significant pest of ornamental and horticultural plants. The salmon louse (Lsl build information, 9,363 clusters, FTP) is an ectoparasitic copepod parasite that can cause significant mortality in farmed and wild salmon. These sets may prove helpful in understanding the biology of parasitism and provide targets for control of these pests.
NCBI Databases in Nucleic Acids Research Database Issue
The Nucleic Acids Research 2011 Database Issue contains nine articles about NCBI resources, tools, and databases including Gene, GEO, Epigenomics, CDD and GenBank. Free full-text articles from the database issue are available from PubMed Central and the publisher’s site and are linked to the summaries and abstracts in PubMed.
dbSNP BLAST Pages Updated
The dbSNP BLAST page has an updated submission form and output format. The new pages have improved organism selection, chromosome specific database selection, and many of the convenient features of the other BLAST services.
New Mammalian Genomes at NCBI
Updated genome annotations for the rat (build 4.2), cow (build 5.2), and a new pig assembly (build 2.1) are now available for searching and viewing in Entrez, BLAST, the Map Viewer, and for downloading from genomes area of the FTP site.
Microbial Genomes Update
Sixty-five finished microbial genomes were released during November and December 2010. The original sequence data files submitted to GenBank/EMBL/DDBJ are available in the Bacteria directory in the genomes area of the GenBank FTP site. RefSeq provisional versions were made for a selected set of 46 these genomes.
In addition, 100 microbial whole genome shotgun-sequencing projects were added to GenBank during this period. The original submitted files are available in the Bacteria_DRAFT directory in the GenBank genomes area. RefSeq provisional versions of 64 of these projects are also available.
All GenBank and RefSeq microbial genomes are incorporated in the NCBI integrated Entrez search and retrieval system.
New Video on NCBI's YouTube Channel
A new video that shows how use My NCBI to save searches and set up automated E-mail alerts for new results is now available on NCBI’s YouTube channel.
GenBank News
GenBank release 181 is available through the NCBI web and FTP sites. The current release incorporates data available as of Dec 15, 2010 and contains 122,082,812,719 bases from 129,902,276 sequence records. Release notes describe the current state of data and upcoming changes.
RefSeq News
RefSeq Release 45 is now available through the Entrez system and can be downloaded from the FTP site. This full release incorporates genomic, transcript, and protein data available as of January 7, 2011 and includes 16,748,646 records from 11,536 different species and strains. The release notes describe changes since the last release. New in this release is the inclusion of additional features present on the corresponding UniProt/Swiss-Prot record for a subset of RefSeq proteins. These new features are indicated with a Note that identifies the source accession number. An example from NP_080213.3 is shown below.
Site 147
/site_type="phosphorylation"
/experiment="experimental evidence, no
additional details recorded"
/note="Phosphoserine; propagated from
UniProtKB/Swiss-Prot(Q9D0F4.1)"
The RefSeq Homepage has more information on the RefSeq project.
Journals Database Now a Part of NLM Catalog
The NCBI Journals Database is now part of NCBI NLM Catalog. The NLM Catalog contains the detailed MEDLINE indexing information for the journals in PubMed and other NCBI databases and will maintain the functions of the Journals database.
Announce Lists and RSS Feeds
Eighteen topic-specific mailing lists are available which provide email announcements about changes and updates to NCBI resources including dbGaP, BLAST, GenBank, and Sequin. The various lists are described on the Announcement List summary page: www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html.
To receive updates on the NCBI News, please see: www.ncbi.nlm.nih.gov/About/news/announce_submit.html.
Twelve RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce. Please see: www.ncbi.nlm.nih.gov/feed/.
Users can also stay updated on NCBI’s resources on Facebook and Twitter: www.twitter.com/NCBI.
Send comments and questions about NCBI resources to vog.hin.mln.ibcn@ofni, or call 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
Publication Details
Author Information and Affiliations
Publication History
Created: January 10, 2011; Last Update: January 28, 2011.
Copyright
Publisher
National Center for Biotechnology Information (US), Bethesda (MD)
NLM Citation
Cooper P, Morris R. NCBI News, January 2011. 2011 Jan 10 [Updated 2011 Jan 28]. In: NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.