NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
How has the SRA grown!
Monday, September 24, 2012

RefSeq Release 55 is out!
Thursday, September 20, 2012
Now in PubChem: >6 million chemicals from SCRIPDB with links to USPTO patents
Thursday, September 20, 2012
More than 6 million structures from SCRIPDB are now available in (PubChem. Extracted from the complex work units of more than 300,000 USPTO patents between the years of 2001-2012, these PubChem Substance records link to both SCRIPDB and the USPTO websites.
Click here to retrieve the SCRIPDB records in PubChem Substance.
A full text article describing the SCRIPDB project is available in PubMed Central.
New CDD release contains 239 new or updated NCBI-curated domains
Wednesday, September 19, 2012
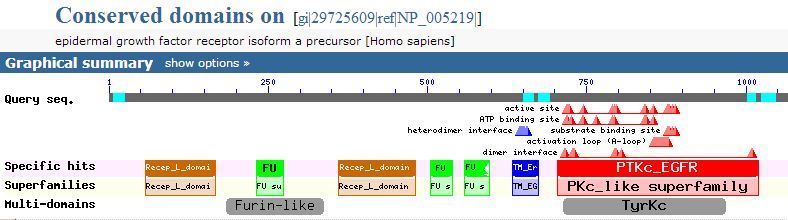
CDD data has been incorporated in the Entrez and BLAST search services and CDD matrices and other information can be downloaded from the CDD FTP site.
PubChem databases and services are now HTTPS compatible
Monday, September 17, 2012
PubChem, NCBI's small molecule database and search system, is now Hypertext Transfer Protocol Secure (HTTPS) compatible, allowing encryption in interactions with the site.
PubChem's PUG REST 1.0 is now available!
Thursday, September 13, 2012
PubChem Power User Gateway (PUG) REST interface 1.0 version is now available and replaces the April beta release!
This service allows access to PubChem data and services through simple HTTP requests, for use in scripts, javascript embedded in web pages, and 3rd party applications, without the overhead of XML and SOAP envelopes that are required for other versions of PUG. Unlike other PubChem services, PUG REST can deliver customized output or specific data elements without having to download the full record and can accept a variety of input types such as chemical name, InChIKey, SMILES in addition to numeric identifiers. The PUG REST help document provides more information on formulation HTTP requests and the available functions. A new PUG REST tutorial provides examples of PUG REST.
The BLAST+ User Manual has been revised & updated
Thursday, September 13, 2012
The BLAST+ manual available on the NCBI Bookshelf has been extensively revised and updated. Appendix C now shows all available options for the different BLAST+ programs.
Stand-alone BLAST has been updated
Wednesday, September 12, 2012
Stand-alone BLAST version 2.2.27+ is now available for download from the BLAST Executables FTP site. This new version contains a number of important improvements and some bug fixes. Improvements include more accurate composition-based statistics for translating searches (blastx), the ability to run remote searches with deltablast, reduced memory usage by blastn with short queries, and improved gap placement by blastn in megablast mode. See the Blast News for more detail.
PubChem reaches milestones on its 8th BDay!
Wednesday, September 12, 2012

A new version of Genome Workbench is available
Thursday, September 06, 2012
A new version (2.6.0) of NCBI's Genome Workbench is now available. Genome Workbench is a standalone sequence viewer, annotation, and analysis platform. This version has many new features, improvements, and a few bug fixes that are described in the Release Notes.
NCBI is now using Genome Annotation Release numbers
Tuesday, September 04, 2012
The NCBI eukaryotic genome annotation pipeline is now using Annotation Release numbers to decrease confusion regarding the independent notions of a genome assembly and its annotation.
Annotation Release numbers:
- are integer values that increment each time the genome annotation is updated.
- have initial values starting at 100 or higher. are incremented independently for each organism.
- are used for the set of annotations calculated on one or more genome assemblies.
Please see the documentation describing The NCBI eukaryotic Genome Annotation Process page for more information.
- How has the SRA grown!
- RefSeq Release 55 is out!
- Now in PubChem: >6 million chemicals from SCRIPDB with links to USPTO patents
- New CDD release contains 239 new or updated NCBI-curated domains
- PubChem databases and services are now HTTPS compatible
- PubChem's PUG REST 1.0 is now available!
- The BLAST+ User Manual has been revised & updated
- Stand-alone BLAST has been updated
- PubChem reaches milestones on its 8th BDay!
- A new version of Genome Workbench is available
- NCBI is now using Genome Annotation Release numbers
- NCBI News, September 2012 - NCBI NewsNCBI News, September 2012 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...
