NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
New Science Feature on NCBI Insights - Transcriptome of Tasmanian devil and its transmissible cancer
Thursday, February 28, 2013
A new Science Feature is now on NCBI Insights Bog - "The Tasmanian Devil and Cancer as an Infectious Disease: Analysis of transcriptome data." Recent research about a strange and deadly infection that causes a transmissible cancer in the Tasmanian devil has provided considerable data and insights on the mechanism of pathology.
This post shows how to access the relevant literature and these RNA-Seq data through PubMed Central, the BioProjects, and Sequence Read Archive (SRA). It also demonstrates using BLAST and the SRA run browser to analyze expression levels of specific protein coding and miRNA genes in these data sets.

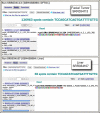
Figure
Figure 1. SRA BLAST results using the devil POMC transcript as a query against testis transcriptome data (top panel) and facial tumor transcriptome data (bottom panel). This gene is highly expressed in the tumor sample.
Figure
Figure 2. Filtering SRA runs for liver (top panel) and facial tumor(bottom panel) using the stem sequence of MIR338 brain-specific miRNA. High counts for this sequence in the tumor run is consistent with a neural origin for this tumor.
New Quick Tip on NCBI Insights Blog - how to download bacterial genomes using the Entrez API
Wednesday, February 20, 2013
A new Quick Tip, "How to Download Bacterial Genomes Using the Entrez API" on the NCBI Insights blog shows how to download bacterial genomes programmatically for a list of species using the E-utilities, the application programming interface (API) to NCBI’s Entrez system of databases.
This strategy takes advantage of NCBI’s redesigned Genome database that links all genome sequences for a given species to one record, making it easy to obtain the desired sequences after finding the right Genome record. The post includes a demonstration script that is easy to adapt to download genomes of interest.
GenBank Release 194.0 is Available
Tuesday, February 19, 2013
The new release for GenBank is now available via ftp.ncbi.nlm.nih.gov, as well as in the Nucleotide database and BLAST services.
In release 194.0 (02/15/2013), the total number of non-WGS, non-CON records was comprised of basepairs of sequence data. In addition, there were 103,101,291 WGS records containing 390,900,990,416 basepairs of sequence data.
During the 63 days between the close dates for GenBank Releases 193.0 and 194.0, the non-WGS/non-CON portion of GenBank grew by 1,750,490,954 basepairs and by 1,746,402 sequence records, with an average of 45,507 non-WGS/non-CON records added and/or updated per day. In addition, the WGS component of GenBank grew by 34,898,067,578 basepairs and by 10,333,526 sequence records.
The total number of sequence data files increased by 45 with this release, with the divisions that expanded in file number:
- BCT = 4 new files, now a total of 98
- CON = 8 new files, now a total of 187
- ENV = 2 new files, now a total of 59
- GSS = 4 new files, now a total of 270
- INV = 1 new file, now a total of 34
- PAT = 4 new files, now a total of 190
- TSA = 5 new files, now a total of 138
- VRT = 2 new files, now a total of 30
The total number of AUT (author name) index files increased by 4 with this release and is now composed of 110 files.
For downloading purposes, please keep in mind that these GenBank flatfiles are roughly 587 GB (sequence files only) or 632GB (including the 'short directory', 'index' and *.txt files) when uncompressed. The ASN.1 formatted datafiles are approximately 481 GB.
For additional release information, see the Release Notes and README files in individual directories.
One change is noteworthy of mentioning here as described in the Release Notes sections 1.3.2 & 1.3.3:
With GenBank 193.0 experimental "Release Catalog" products were produced in a plan to eventually replace the old GenBank "index" files. GenBank release 193.0 was the last release for which the legacy index files will be provided.
Shortly, Release Catalogs and other supplemental files for Release 194.0 will be made available. In these files every GenBank sequence record will be represented by a 10-field, TAB-delimited row of data. [Please note that when a value does not exist for one of these fields, the field in the catalog will be empty (eg, two sequential TAB characters can be present, with nothing between them.]
The new files will be made available in a new sub-directory of the GenBank FTP area: ftp.ncbi.nlm.nih.gov/genbank/catalog.
The data fields in the Release Catalog files are:
- Accession Number
- Accession.Version
- NCBI GI Identifier (if assigned)
- Molecule Type (dna, rna, mrna, etc)
- Sequence Length
- Organism Name
- NCBI Taxonomy Database Identifier
- Division Code
- BioProject Accession Number
- BioSample Accession Number
Here is an example of a row of Release Catalog data for the entry CP003933:
- CP003933 CP003933.1 429549985 dna 3618794 Sinorhizobium meliloti GR4 1235461 BCT PRJNA175860
In addition, new "PMID List" and "Gene List" TAB-delimited files will accompany the Release Catalog.
The format of the PubMed Identifier List file is:
- Accession1 Accession1.Version PMID-1,PMID-2,PMID-3,.....
- Accession2 Accession2.Version PMID-1,PMID-2,PMID-3,.....
- ....
The format of the Gene List file is:
- Accession1 Accession1.Version Gene-Symbol-1 Locus-Tag-1
- Accession1 Accession1.Version Gene-Symbol-2 Locus-Tag-2
- ....
We plan to provide the release catalog and accompanying lists via files that are specific to EST, GSS, and non-EST/GSS (everything else), however at least initially the catalogs and lists will not include the contig sequence records for WGS projects.
NCBI Insights' First Quick Tip: How to find functional protein homologs using conserved domains
Tuesday, February 12, 2013
Protein researchers often want find homologous proteins in different species. In the first "Quick Tips & Tricks" post of the NCBI Insights Blog, "Using Conserved Domains to Find Functional Homologs" describes a step-by-step method for how to do this using curated functional domain information and links provided in the Conserved Domains Database (CDD).
New NCBI Insights Blog Explains the IE7 Warning
Tuesday, February 05, 2013
For many months, people viewing NCBI webpages using the web browser Internet Explorer 7 have seen a warning on the top of their webpages. A new NCBI Insights Blog explains to users what the Internet Explorer 7 warning means.
The NCBI Insights blog post explains:
- Why the IE7 web browser is no longer supported.
- What “stop supporting this browser” means.
- What actually happened on the NCBI website on January 1, 2013.
- What this really means about for people using IE7 to view the NCBI website.
Links are provided for more information and to the most up-to-date web browsers.
Click here to read the article: "What does NCBI’s Internet Explorer 7 warning mean?".
PubReader Article View Now In Use By KoreaMed Synapse
Friday, February 01, 2013
KoreaMed Synapse (http://synapse.koreamed.org), a digital archive and reference linking platform of Korean medical journals, is now using NCBI’s new PubReader presentation style to display their full-text journal articles.
KoreaMed’s database of over 120 journals now includes a blue ‘PubReader’ icon for each full-text article. NCBI launched PubReader in December 2012 as a convenient new way to view full-text articles in PubMed Central on desktops as well as tablets and mobile devices. In tandem with the launch, NCBI made the code used to create PubReader freely available on GitHub at https://github.com/NCBITools/PubReader.
- New Science Feature on NCBI Insights - Transcriptome of Tasmanian devil and its transmissible cancer
- New Quick Tip on NCBI Insights Blog - how to download bacterial genomes using the Entrez API
- GenBank Release 194.0 is Available
- NCBI Insights' First Quick Tip: How to find functional protein homologs using conserved domains
- New NCBI Insights Blog Explains the IE7 Warning
- PubReader Article View Now In Use By KoreaMed Synapse
- NCBI News, February 2013 - NCBI NewsNCBI News, February 2013 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...