NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
New publication: "BLAST: a more efficient report with usability improvements."
Tuesday, April 30, 2013
A new publication, "BLAST: a more efficient report with usability improvements," (PMID: 23609542) is now available in free full-text from the Webserver Issue of Nucleic Acids Research. The paper describes the recent improvements in the NCBI BLAST Web output. These include more efficient loading of results, the ability to retrieve only the aligned regions, to display query-based or subject-based views of results in the graphical sequence viewer and to customize the descriptions table. A factsheet and a video on the NCBI YouTube channel provide a practical introduction to these features.
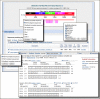
Figure
The re-designed BLAST results showing many of the new options.
"A Librarian's Guide to NCBI" Course was a Success!
Monday, April 29, 2013
Last week (April 15-19, 2013), NCBI in collaboration with the National Library of Medicine and the National Network of Libraries of Medicine NLM Training Center at the University of Utah presented "A Librarian’s Guide to NCBI". This new course, which was highly rated by participants, was designed to prepare health science librarians for supporting and training patrons about NCBI molecular databases and tools at their own institutions.
As promised in last week’s NCBI Insights Blog post, the materials used in "A Librarian's Guide to NCBI" are now available for download and use for personal enlightenment or to supplement training in workshops or courses.
On a typical day the course offered two modules, each one focused on a different aspect of molecular data at NCBI and included a short lecture followed by an assessment quiz, instructor-led practical demonstrations, and individual practice problems. In addition to the modules, there were two discussion sessions reviewing library patron questions provided by participants, an open question and answer session with NCBI engineering branch supervisors, a tour of the National Library of Medicine, and a visit by NCBI Director David Lipman. An online forum for the librarian cohort is being developed for continued communications and support.
Based on strong participant evaluations and requests, we are planning to offer the Librarian’s Guide at least once a year.
Check back on NCBI’s Education page for future offerings of this and other NCBI courses.
____________________________________________
In addition to their usage in "A Librarian's Guide to NCBI", the curricular materials were developed as separate stand-alone modules to be used by educators and bioinformatics trainers. These are now available for download on the Librarian Course FTP site.
Modules were developed to explain and demonstrate related NCBI resources for use by researchers of broad biology-based disciplines. In each of the eight modules the following questions were answered:
- Why are the data generated?
- How are the data generated / determined / measured?
- How does the NCBI organize and represent the data?
- What tools are available at the NCBI to analyze / search the data?
- What experimental questions can be answered with the data?
- What are the caveats / limits of data interpretation?
- What would library patrons want to do with the data?
Each module featured a 30-minute lecture followed by a brief assessment quiz with a discussion of the answers, instructor-led practical demonstrations, and individual practice problems, and topics covered were:

Figure
Sample slides from the eight modules of A Librarian’s Guide to NCBI. Complete PowerPoint files are available from the Librarian Course FTP site.
- Molecular Biology Basics - a review of molecular biology concepts focusing on biological information flow and the gene as a central theme and showed how the NCBI Gene database serves as a central access point for molecular data at NCBI.
- Advanced Entrez Searching - a demonstration of how to use the Entrez integrated database and search system to find relevant data using both basic and advanced interfaces and fielded searches. The module also demonstrated the importance of pre-compiled and pre-computed relationships for navigating within a database and laterally across the Entrez system.
- NCBI BLAST - a full-day introduction to sequence similarity searching using NCBI’s Basic Local Alignment Search Tool (BLAST). This module covered the basics of sequence alignment algorithms, scoring matrices, and local alignment statistics and used practical protein and nucleotide search examples that highlighted features of the BLAST web service designed to give the most relevant results.
- Sequences & Genomes - an exploration of the essential role of nucleotide and protein sequence data in modern biological research and the Nucleotide database as the backbone of the NCBI molecular databases. The module explained how NCBI manages and processes sequence and other data associated with genomes and their annotation. Demonstrations and exercises showed how to identify the most up-to-date and well-annotated sequence.
- Sequence Variation and its Consequences - an examination of the many databases and tools at NCBI that provide access to variation data emphasizing the association between variation and disease risk. After describing the different types of genetic variation as well as the major study methods that produce these data, practical demonstrations and exercises demonstrated how to navigate the NCBI variation resources to find specific data and important attributes, such as geographic population, allele frequency, and disease association.
- Gene Expression & Biological Pathways - a review of NCBI databases and tools relevant to the study of gene expression. The module provided basic background on the importance of gene expression in various biological phenomena and high-throughput techniques for measuring expression. Practical demonstrations showed how to find and compare expression patterns of genes in different samples in microarray datasets and expression profiles, and how to map selected genes onto metabolic pathways.
- Protein Structures - an illustation of the usefulness and interconnectedness of NCBI protein structure databases and tools using the example DNA Topoisomerase II. The module covered basic concepts of structural biology and the importance of 3D structure information in understanding the normal functions of proteins and abnormal functions that result in disease. Practical examples showed how to find available 3D structural data for a given protein sequence, detect functional domains within the sequence, view 3D structure data using Cn3D, and explore the relationship between protein sequence and structure data.
- Drugs & Other Small Molecules - a tour of NCBI’s Chemical and Bioactivity Databases developed by The PubChem Project. The module explained and explored the data in and relationship between PubChem databases (Compound, Substance and BioAssay). Practical examples elucidated the types of data that are accessible from these resources, and provided case-study specific, guided demonstrations for finding information to answer important scientific questions.
Based on course feedback, we plan to expand the course materials to include a set of videos of the lectures and demonstrations to be produced for the NCBI YouTube Channel as well as a set of worked exercises suitable for classroom teaching.
Visit NCBI’s Education page for links to these and other training materials.
GenBank Release 195.0 is Available
Tuesday, April 16, 2013
The new release for GenBank is now available via ftp.ncbi.nlm.nih.gov, as well as in the Nucleotide database and BLAST services.
In release 195.0 (04/11/2013), the total number of non-WGS, non-CON records was comprised of basepairs of sequence data. In addition, there were 164,136,731 WGS records containing 151,178,979,155 basepairs of sequence data.
During the 57 days between the close dates for GenBank Releases 194.0 and 195.0, the non-WGS/non-CON portion of GenBank grew by 1,037,624,297 basepairs and by 1,250,004 sequence records, with an average of 32,964 non-WGS/non-CON records added and/or updated per day. In addition, the WGS component of GenBank grew by grew by 27,125,603,190 basepairs and by 7,408,023 sequence records.
The total number of sequence data files increased by 30 with this release, with the divisions that expanded in file number:
- BCT = 2 new files, now a total of 100
- CON = 8 new files, now a total of 205
- ENV = 1 new file, now a total of 60
- EST = 3 new files, now a total of 472
- PHG = 1 new file, now a total of 2
- PLN = 1 new file, now a total of 61
- TSA = 3 new files, now a total of 141
- VRL = 1 new file, now a total of 25
For downloading purposes, please keep in mind that these GenBank flatfiles are roughly 594 GB (sequence files only).
For additional release information, see the Release Notes and README files in individual directories.
New Educational Initiative: A Librarian's Guide to NCBI
Thursday, April 11, 2013
Next week NCBI will premiere A Librarian’s Guide to NCBI, a new course aimed at teaching health science librarians about NCBI resources. For more information, a new NCBI Insights Blog introduces the course and updates on the course and the availability of the curricular materials will be publicized on Twitter and Facebook.
A Librarian’s Guide to NCBI Course Modules
| Day 1 |
 |
| Day 2 |
 |
| Day 3 |
 |
| Day 4 |
 |
| Day 5 |
 |
PubChem Releases New and Enhanced Webpage Widgets
Wednesday, April 10, 2013
New PubChem Widgets (Chemical Structure Carousel, Classification Listing and Autocomplete) have been developed for you to use in your own webpage. In addition, existing table-based Widgets (including Bioactivity, Patents and PubMed) have been enhanced with a Link/Embed button that allows you to open the widget in a PubChem page or embed the widget in your own page as an iframe.
- The Structure Carousel displays chemical structure thumbnail images along with names/synonyms, and will also show related annotations, when available, such as medication information, literature, patents, bioactivities, and 3D structures.
- The Classification Listing displays the classification, when available, of a PubChem Compound, Substance, or BioAssay. Current Classifications include MeSH, ChEBI, KEGG, LIPID MAPS, and Gene Ontology.
- The Autocomplete Widget is an embeddable tool that suggests a list of terms when you type input into a search field.
For complete documentation about all PubChem Widgets, see: http://pubchem.ncbi.nlm.nih.gov/widget/docs/widget_help.html
BLAST 2.2.28 now available
Friday, April 05, 2013
Stand-alone BLAST version 2.2.28+ is now available for download from the FTP site. BLAST 2.2.28+ provides a number of important new features, improvements and some bug fixes. New features include composition-based statistics for Reverse PSI-BLAST (rpsblast), expanded options (query coverage, subject title, and taxonomy) for tabular output, and batch subsequence retrieval in blastdbcmd. Improvements include adaptive BATCH_SIZE resulting in more efficient searching, and incremental production of XML results. The Blast Release Notes have more details.
Try it out! The New PubChem Upload Beta Site
Friday, April 05, 2013
A new beta version PubChem Upload system is available to try out. It features streamlined procedures for data submissions and updates to both the PubChem Substance and BioAssay databases.
The new capabilities offered by PubChem Upload include:
- Assay & Substance wizards to assist novice users
- Greatly improved UI speed using newer web technology (minimizing possible time-outs)
- Easy new user registration/easy upgrade
- Improved help with a tutorial and hints built into user interface
- Substance input in varied formats CID, SID, SMILES, etc.
- PubChem substance/assay templates for new submissions or for record updates
- Error display integrated with substance list displays
- Full editing and integration of assay data & description tables
- Expanded import/export for data description table spreadsheets
This system will eventually replace the original Pubchem Deposition Gateway.
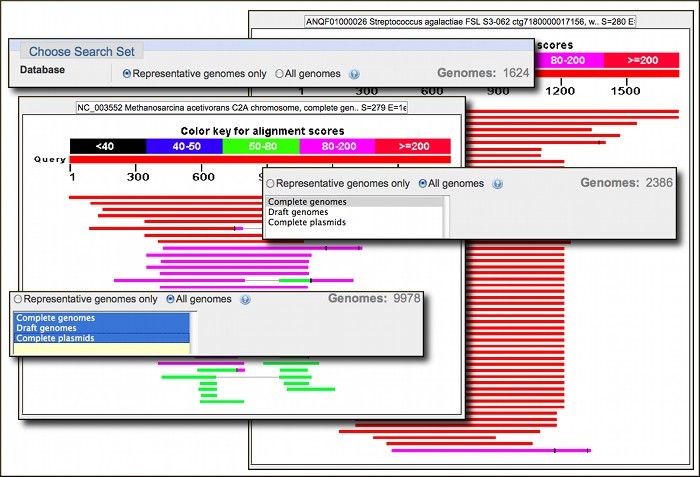
New database options in Microbial Genomes BLAST: Representative Genomes
Friday, April 05, 2013
Microbial Genomes BLAST has new database options including 'Representative genomes', now the default database, and 'All genomes'. Representative genomes provide a smaller less redundant set of records for a given bacterial species. These representatives are selected by the research community and NCBI computational processes and are especially helpful for microbial species that are highly represented by genomes for numerous strains in NCBI databases, such as Escherichia coli. The 'All genomes' option offers the choice of Complete genomes, Draft genomes, or Complete plasmids. You can search these sets individually or in any combination. The microbial BLAST report also has a new 'Genome' link to the species page in Entrez Genome in the alignments section of the BLAST report. Run a search.
- New publication: "BLAST: a more efficient report with usability improvements."
- "A Librarian's Guide to NCBI" Course was a Success!
- GenBank Release 195.0 is Available
- New Educational Initiative: A Librarian's Guide to NCBI
- PubChem Releases New and Enhanced Webpage Widgets
- BLAST 2.2.28 now available
- Try it out! The New PubChem Upload Beta Site
- New database options in Microbial Genomes BLAST: Representative Genomes
- NCBI News, April 2013 - NCBI NewsNCBI News, April 2013 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...

