NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Come to the NCBI Discovery Workshops on July 30 & 31!
Friday, June 28, 2013
Spaces are still available for the free, 2-day NCBI Discovery Workshops to be held on the NIH Campus on July 30 and 31, 2013. For more information and to register, visit the Discovery Workshops homepage.
The NCBI Discovery Workshops comprise four workshops that will teach you how to use the NCBI Web resources more effectively. The July 2013 Workshops consist of four 2.5-hour hands-on sessions, with each session focusing on a different related group of NCBI tools and databases:
Materials from all Discovery Workshops offerings are available from the Education FTP directory.
Upload and graphically compare your own data with NCBI Epigenomics tracks
Wednesday, June 26, 2013
Recently, a new “Upload Tracks” system has been added to the NCBI Epigenomics resource to allow users to view and compare their own data with information stored at NCBI.
The NCBI Epigenomics resource, a comprehensive public repository for whole-genome epigenetic datasets, contains information from a subset of data in the Gene Expression Omnibus (GEO), which has been subjected to additional review and annotation. Currently there are over 4200 viewable and downloadable datasets from over 1200 samples that have been isolated from five well-studied species.
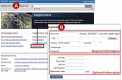
From the NCBI Epigenomics homepage (Figure 1A), you can access the “Upload” page where your own datasets can be uploaded and displayed as tracks in the Epigenomics genome viewer.
Please note that the “Upload Tracks” feature requires logging into a My NCBI Account which facilitates the storing of information for future examination and also ensures that the data is only visible and viewable by the account holder. The uploaded information cannot be viewed, downloaded or used by any other user of the Epigenomics resource.
Once logged into NCBI, the Epigenomics “Upload” page (Figure 1B) contains fields for entering important metadata information as well as the dataset itself into the system.
Each user-uploaded dataset is listed in the “My Uploads” collection as an experiment (Figure 2A). This allows for uploaded data tracks to be selected for operations such as adding to user-created collections or viewing in concert with other database tracks in the customizable genome viewer interface (Figure 2B).
FOR MORE INFORMATION:
Epigenomics Homepage & Epigenomics Upload Page
- Uploading Epigenomics DataSets - YouTube Video | HelpDoc
- Using the Epigenomics Genome Viewer - YouTube Video | HelpDoc
My NCBI - Sign In/Registration Page & YouTube Video
SRA-BLAST has been updated with new features and performance enhancements
Tuesday, June 25, 2013
SRA-BLAST has undergone a dramatic update, both in terms of user interface and search performance. These updates to SRA-BLAST make it an even more useful tool for searching through more than 700 trillion open-access bases currently housed within the Sequence Read Archive (SRA).
New features include:
- Targeted searching within one or more SRA Experiment sets (i.e., "SRX accessions"). Users may now search combined datasets of up to 2 billion individual reads.
- An "autocomplete" feature that will allow users to specify SRX accession, SRX title, organism scientific name, or tax id to help build the search set.
- Data obtained from Roche 454 and newer Illumina instruments (HiSeq and MiSeq).
Welcome to the NCBI News site!
Tuesday, June 25, 2013
This is the place to get the latest information about NCBI, and feature stories about NCBI services and activities. The NCBI News site offers readers fast and integrated access to the most important news stories about announcements, changes, updates and improvements at NCBI.
This site replaces the NCBI Newsletter, previously published on the NCBI Bookshelf. In comparison to the Bookshelf Newsletter, News stories will have a more rapid publication cycle enabling the release of announcements and updates as they become available.
In the “Follow us” portlet, the News site displays icons which link directly to NCBI’s RSS Feeds and Email ListServs in a single, easily accessible place, as well as icons which link to the NCBI Insights Blog and NCBI’s social media outlets on Twitter , Facebook, and YouTube.
In addition to reading articles describing changes and updates to our website and data, the NCBI News site offers the ability to share interesting information with others. By clicking the “Share” button located at the top and bottom of each news story, readers can post the title of the article and a link directly to social media sites such as Facebook, Twitter, LinkedIn, WordPress, Reddit, Tumblr, Pinterest, StumbleUpon, and many others....
Dr. David Lipman Receives White House "Open Science" Champions of Change Award on Behalf of NCBI
Thursday, June 20, 2013
Dr. David Lipman, Director of the NLM’s National Center for Biotechnology Information (NCBI), was among those honored by the White House on June 20 for their outstanding work in “promoting and using open scientific data and publications to accelerate progress and improve our world” as a White House "Open Science" Champion of Change.
As Director of the NCBI, Dr. Lipman was honored for his leadership in making biomedical data and health information publicly and easily available to all, including scientists, medical professionals, patients, educators and students.
“I am truly honored that the White House has recognized our work in providing resources such as NCBI’s GenBank database of all publicly available DNA sequences and PubMed Central, an online archive of peer-reviewed biomedical sciences literature,” said Dr. Lipman. “The success of these databases and NCBI’s many other resources is a reflection of the hard work, dedication and talent of all those working at NCBI.”
GenBank Release 196.0 is Available
Tuesday, June 18, 2013
The new release for GenBank is now available via ftp.ncbi.nlm.nih.gov, as well as in the Nucleotide database and BLAST services.
Release 196.0 (06/13/2013) 165,740,164 non-WGS, non-CON records which were comprised of 152,599,230,112 basepairs of sequence data. In addition, there were 112,488,036 WGS records containing 453,829,752,320 basepairs of sequence data.
During the 63 days between the close dates for GenBank Releases 195.0 and 196.0, the non-WGS/non-CON portion of GenBank grew by 1,420,250,957 basepairs and by 1,603,433 sequence records. During that same period, 590,119 records were updated. An average of 34,818 non-WGS/non-CON records were added and/or updated per day. In addition, between releases 195.0 and 196.0, the WGS component of GenBank grew by 35,803,158,714 basepairs and by 1,978,722 sequence records.
The total number of sequence data files increased by 19 with this release, with the divisions that expanded in file number:
- BCT = 3 new files, now a total of 103
- CON = 3 new files, now a total of 208
- ENV = 1 new file, now a total of 61
- EST = 1 new file, now a total of 473
- GSS = 3 new files, now a total of 273
- INV = 1 new file, now a total of 35
- PAT = 5 new files, now a total of 195
- PLN = 1 new file, now a total of 62
- ROD = 1 new file, now a total of 31
For downloading purposes, please keep in mind that these GenBank flatfiles are roughly 600 GB (sequence files only).
For additional release information, see the Release Notes and README files in individual directories.
New RefSeq Bacterial Protein Products and Emerging RefSeq Data Model
Tuesday, June 11, 2013
The NCBI Reference Sequence Project (RefSeq) project is now producing a non-redundant set of sequences to serve as annotation reagents for bacterial genomes. This is to help reduce and control redundancy in the protein databases while maintaining information content in response to high volume sequencing and annotation of multiple isolates. These new protein records begin with the accession prefix ‘WP’ and are used represent each unique bacterial sequence in the RefSeq data. These new proteins are independent of any particular bacterial genome and can be associated with more than one isolate, strain or species. Bacterial genomes will now be annotated using these WP proteins. Existing RefSeq bacterial sequences (YP and NP) accessions now point to the corresponding WP record, and WP records have replaced ZP accessions, which were formerly annotated on partially assembled whole genome shotgun genomes. Please see the Reference Sequence Announcement for further details and the plan for phasing in the implementation.

Figure
A non-redundant record (WP_000161708) representing the invasion-associated secreted protein SopE from Salmonella enterica.
- Come to the NCBI Discovery Workshops on July 30 & 31!
- Upload and graphically compare your own data with NCBI Epigenomics tracks
- SRA-BLAST has been updated with new features and performance enhancements
- Welcome to the NCBI News site!
- Dr. David Lipman Receives White House "Open Science" Champions of Change Award on Behalf of NCBI
- GenBank Release 196.0 is Available
- New RefSeq Bacterial Protein Products and Emerging RefSeq Data Model
- NCBI News, June 2013 - NCBI NewsNCBI News, June 2013 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...