NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Tenth Anniversary of RefSeq FTP Releases
Friday, July 26, 2013
The July 2013 RefSeq FTP release marks the 10th anniversary of RefSeq comprehensive FTP releases. We mark this occasion with a sincere "Thank you!" to the scientific community for continued interest and support, comments, and useful suggestions for improvements that have been made over the past years.
There has been significant total growth since the first release in June 2003! So, we thought you might be interested in seeing how much the RefSeq data has grown.
Growth in the number of accessions, by molecule type:
|
Type of Sequence | June 2003 (Release 1) | July 2013 (Release 60) | Percentage Growth over 10 years |
|---|---|---|---|
| Genomic | 64,729 | 4,165,752 | 6,336% |
| RNA | 211,803 | 4,243,209 | 1,903% |
| Protein | 785,143 | 32,504,738 | 4,040% |
Growth in the number of species, per node:
| Taxonomic Node | June 2003 (Release 1) | July 2013 (Release 60) | Percentage Growth over 10 years |
|---|---|---|---|
| Complete | 2005 | 28,560 | 1,324% |
| Fungi | 27 | 785 | 2,807% |
| Invertebrates | 80 | 1,121 | 1,310% |
| Microbes | 334 | 20,213 | 5,952% |
| Mitochondria | 417 | 3,793 | 810% |
| Plants | 30 | 349 | 1,063% |
| Plasmids | 36 | 1,501 | 4,069% |
| Plastids | 31 | 359 | 1,058% |
| Protozoa | 39 | 179 | 359% |
| Mammals | 74 | 580 | 684% |
| Non-mammalian Vertebrates | 206 | 1,796 | 772% |
| Viruses | 1179 | 3,536 | 200% |
RefSeq Release 60 is Available for FTP
Friday, July 26, 2013
The complete RefSeq release 60 contains 40,913,699 records, 32,504,738 proteins, 4,243,209 RNAs, and sequences from 28,560 different organisms. See the Release statistics file or Release notes for more information.
There are several important announcements for RefSeq release 60.
Selected announcements described below include:
- A new bacterial protein data model and accession series
- Suppression of some bacterial genomes
- Changes in annotation of human and vertebrate transcript records
- Policy change to allow a mixture of known and model accessions for eukaryotic genes
Please see the release note announcement for RefSeq release 60 and documents in the new announcement directory for the full set of announcements with detailed information.
Bacterial genomes, new protein data model and accession series (WP)
NCBI continues to expand the RefSeq bacterial genomes node to include ALL complete and draft genomes that meet minimum assembly and annotation quality criteria. This means that RefSeq will include more than one genome of the same strain which may be provided through strain population sampling or sequencing to monitor a disease outbreak. NCBI is in the process of re-annotating all bacterial genomes, with the exception of a small umber for which annotation is provided by, or in collaboration with, another group (such as E. coli str. K12 substr. MG1655).
Due to the expanded scope of the RefSeq bacterial node, we anticipated a very large increase in the number of identical (redundant) proteins; therefore, we have introduced a new data model for bacterial proteins whereby we are providing a true non-redundant protein dataset associated with a new accession prefix,'WP'. Details about the new data model with examples was announced between release cycles.
This release includes a new supplemental file providing mapping of WP accessions to tax_id and species name, for the subset of WP accessions that are annotated on genomes of different species. For example, see WP_000002243.1. The mapping file is available in the release-catalog directory.
We strongly encourage you to read the full announcement.
Supression of some bacterial genomes
Please note that some RefSeq bacterial genomes were recently suppressed. This includes unannotated genomes that had not been processed by NCBI's annotation pipeline yet and annotated genomes with identified annotation quality issues. This has resulted in a net decrease in RefSeq bacterial genomic accessions in this release. Many of the suppressed accessions will be reinstated when annotation is provided.
Changes in annotation of human and vertebrate transcript records
Recent changes to human and other vertebrate transcript records includes:
- removal of exon numbers
- expanded reporting of support evidence, in a structured comment with the header 'Evidence Data'
- (new) reporting gene and transcript attributes, in a structured comment with the header 'RefSeq Attributes'
- removal of mitochondrial localization information from the record DEFINITION line (moved to Attributes)
Please see the detailed description of these changes.
Policy change to allow a mixture of known and model accessions for eukaryotic genes
Previously, we did not allow a mixture of X* series accessions (genome annotation models) and N* series accessions (based on cDNA and curation) for a gene. We have changed this policy in order to provide increased annotation of splice variants. RefSeq models are calculated using cDNA, protein, and RNAseq data. There may be good support at the level of each exon pair; however, the long range exon combination represented in the model may not be fully supported and thus is less likely to be represented with a N* series accession. For example, see Gene ID: 100306968.
New NCBI Insights Post: "New Pandoravirus Sequences are Accessible in GenBank"
Wednesday, July 24, 2013
A new NCBI Insights Blog post provides information on a recent article that describes the discovery and characterization of two “giant” viruses that are proposed to comprise the first members of the “Pandoravirus” genus. The authors of this publication have submitted assembled and annotated genomes to NCBI, which are currently available in the Nucleotide database with the accessions KC977571 and KC977570.
For more information see:
- NCBI Insights Blog Post: "New Pandoravirus Sequences are Accessible in GenBank"
- Philippe, et al. "Pandoraviruses: Amoeba Viruses with Genomes Up to 2.5 Mb Reaching That of Parasitic Eukaryotes." Science. 2013 Jul 19;341(6143):281-6. doi: 10.1126/science.1239181.
Genome Workbench 7.6 with Publication Quality Graphics Export
Monday, July 08, 2013
The latest release 2.7.6 (2.7.5) of Genome Workbench, NCBI's standalone sequence analysis and annotation platform, now produces publication quality graphical output (PDF). A new tutorial shows how to use this helpful feature. The release notes have more information on this and other improvements.
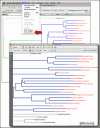
Figure
Exporting a phylogenetic tree view as a PDF from Genome Workbench. The bottom panel shows the high quality PDF.
- NCBI News, July 2013 - NCBI NewsNCBI News, July 2013 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...

