NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Human CCDS release 15 now available on web and FTP
Monday, January 27, 2014
The Consensus Coding Sequence (CCDS) update for Homo sapiens annotation release 105 is now available on the CCDS website and FTP site. This release adds 349 new CCDS IDs to the human CCDS dataset and is based on comparative analysis of NCBI Homo sapiens annotation release 105 and Ensembl release 74. The human CCDS dataset now includes 29,045 proteins that correspond to 18,683 genes.
This is the final CCDS update for human that is based on the human reference assembly GRCh37. The next CCDS update for human will be based on the updated assembly GRCh38 and is tentatively expected to be released in July 2014.
RefSeq release 63 now available
Tuesday, January 21, 2014
The full RefSeq release 63 is now available with nearly 50 million records describing 37,371,278 proteins, 5,760,653 RNAs, and sequences from 33,485 different organisms.
Some important updates include the following:
Directory name change: The RefSeq release directory “microbial” will be removed. Two new directories, “archaea” and “bacteria” will be added. This change will appear in release 65 in May 2014.
WGS process flow change: WGS accessions will no longer be processed on a per-project (WGS prefix) basis. Instead, these accessions will be processed and packaged the same as non-WGS accessions. This will significantly reduce the number of files in the /complete/ and (new) /archaea/ and /bacteria/ directories. Therefore, there will no longer be a series of files named like “microbialNZ_*”. Instead, all WGS scaffolds will be found in concatenated files just like all other accession data. We will continue to provide a separate file for the WGS master records. This change will appear in release 65 in May 2014.
Human Genome GRCh38 Annotation plans: The Genome Reference Consortium released an updated assembly for the human reference genome (GRCh38) in late December 2013. NCBI annotation of the RefSeq copy of this assembly is currently in progress. We anticipate releasing annotation in early to mid-February and including it in RefSeq release 64 in March 2014.
More details about RefSeq release 63 is included in the release statistics and release notes. In addition, reports indicating the accessions included in the release and the files installed are available.
Taxonomy database now shows type material, sequences from type specimens and strains now labeled in Entrez
Tuesday, January 21, 2014
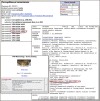
The naming, classification and identification of organisms traditionally relies on the concept of type material, which defines the representative examples ("name-bearing") of a species. For larger organisms, the type material is often a preserved specimen in a museum drawer, but the type concept also extends to type bacterial strains as cultures deposited in a culture collection. Of course, modern taxonomy also relies on molecular sequence information to define species. In many cases, sequence information is available for type specimens and strains. Accordingly, the NCBI has started to curate type material from the Taxonomy database, and are using this data to label sequences from type specimens or strains in the sequence databases. The figure below shows type material as it appears in the NCBI taxonomy entry and a sequence record for the recently described African monkey species, Cercopithecus lomamiensis.
Sequence from type material is particularly important because the species identification is virtually certain to be correct. The Entrez query "sequence from type"[filter] can be used to retrieve these sequence entries and can be used in combination with other queries as in the following examples.
By Organism
By Collection
- "sequence from type"[filter] AND collection cbs (type strains at the CBS culture collection)
- "sequence from type"[filter] AND collection mcz (type specimen at the Museum of Comparative Zoology)
By Author
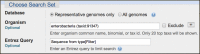
As shown in the figure below, “sequence from type”[filter] is also useful as an Entrez query to limit BLAST searches to reliably identified sequences, particularly when working with prokaryotes.
Stay tuned for more developments and added features coming to the Taxonomy database.
New NCBI Insights blog: NCBI Remap tool helps you transition to newest human reference genome assembly, GRCh38
Thursday, January 16, 2014
NCBI’s Genome Remapping Service (NCBI Remap) allows you to map annotation data from one genomic assembly to another for a selected set of organisms. This may be particularly helpful in updating your annotations for the human reference genome assembly, which has recently been updated to the new version, GRCh38. The newest blog post on NCBI Insights describes how Remap works and how it allows you to analyze data in the context of the newest genome assembly. Finally, the blog post provides links to Remap-related documentation including an overview, FAQs, and a YouTube tutorial.
Sequence Viewer PDF rendering available - YouTube video tutorial
Tuesday, January 14, 2014
NCBI has created a YouTube video tutorial showing you how to generate a PDF rendering of your Sequence Viewer display. The short clip takes you through the downloading process, and shows you what you can do with your file after creating a PDF.
Genome Workbench Update 2.7.12
Tuesday, January 14, 2014
Genome Workbench 2.7.12 has been released. The update includes several new features like a faster Tree Renderer, redesigned data import and an improved GFF format reader. The release notes include more information on features, fixes and improvements.
VAST+ released: Find similar 3D structures for macromolecular complexes
Thursday, January 09, 2014
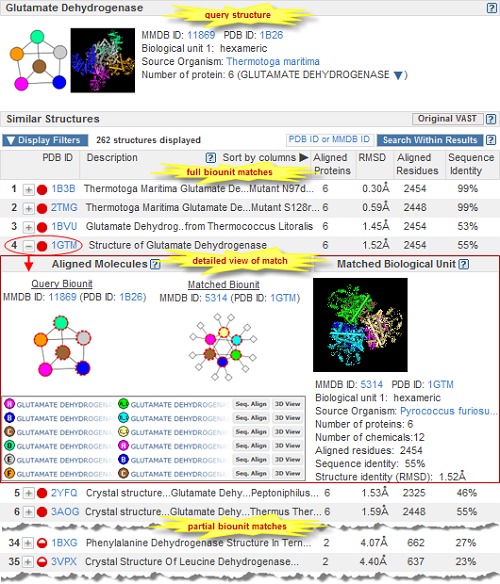
VAST+ is a new tool designed to identify macromolecules that have similar 3-dimensional structures with an emphasis on finding similar macromolecular complexes. The similarities are calculated using purely geometric criteria without regard to sequence similarity, and therefore can identify distant homologs.
This new tool is built upon the original Vector Alignment Search Tool (VAST) and expands the capabilities of that program by taking into account the biological unit ("biounit") of each structure, not just individual protein chains or their substructures.
A recent publication provides detailed information about the VAST+ algorithm. In addition, the extensive VAST+ help document includes a comparison of original VAST and VAST+, as well as examples of how can this tool can be used to learn more about proteins.
Please note that in order to view the 3D superpositions of similar biological units, you must install the most recent version of the NCBI molecular viewing software, Cn3D 4.3.1.
Figure. VAST+ search results for Thermotoga maritima Glutamate Dehydrogenase (PDB ID 1B26) with detailed view of a match to 1GTM.
For more information:
NCBI Insights blog: A Librarian's Guide to NCBI - an intensive training course for medical librarians to be offered April 2014
Wednesday, January 08, 2014
The NCBI in partnership with the National Library of Medicine Training Center (NTC) will offer the Librarian’s Guide to NCBI course on the NIH campus in April 2014. This will be the second presentation of the course; it was previously offered in the spring of 2013 (NCBI Insights April 11 and May 6, 2013). After the course, we will post lecture slides and hands-on practical exercises on the education area of the NCBI FTP site and video tutorials of the course lectures will be available on the NCBI YouTube channel. Materials from the 2013 course are available, as well as lecture videos for the expression module. More information, including prerequisites, is available in the newest NCBI Insights blog post.
Mouse genome annotation release 104 available
Wednesday, January 08, 2014
The mouse (Mus musculus) genome annotation has recently been updated to annotation release 104 and is now available in the Nucleotide, Protein sequence and Gene databases, is searchable using BLAST, and can be downloaded from the FTP site.
Mouse annotation release 104, based on the sequence assemblies GRCm38.p2 (GCF_000001635.22) and Mm_Celera (GCF_000002165.2), identifies a total of 35,389 genes, as well as 100,581 transcripts on GRCm38.p2. RNA-Seq data from 31 distinct BioSample accessions were aligned to assist in gene prediction. More statistics are available in the annotation report.
See what other annotation runs are in progress on the Eukaryotic Genome Annotation Pipeline status page.
BLAST+ 2.2.29 now available
Tuesday, January 07, 2014
Stand-alone BLAST version 2.2.29+ is now available for download from the FTP site. BLAST 2.2.29+ provides a number of important improvements and bug fixes. Some improvements include improved blastn batch query performance, source releases build optimized multi-thread binaries by default, and improved multithreading by better dividing the BLAST database among threads. The BLAST Release notes lists more upgrades and fixes.
- Human CCDS release 15 now available on web and FTP
- RefSeq release 63 now available
- Taxonomy database now shows type material, sequences from type specimens and strains now labeled in Entrez
- New NCBI Insights blog: NCBI Remap tool helps you transition to newest human reference genome assembly, GRCh38
- Sequence Viewer PDF rendering available - YouTube video tutorial
- Genome Workbench Update 2.7.12
- VAST+ released: Find similar 3D structures for macromolecular complexes
- NCBI Insights blog: A Librarian's Guide to NCBI - an intensive training course for medical librarians to be offered April 2014
- Mouse genome annotation release 104 available
- BLAST+ 2.2.29 now available
- NCBI News, January 2014 - NCBI NewsNCBI News, January 2014 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...