NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Genome Workbench Update 2.7.15 released
Wednesday, February 26, 2014
Genome Workbench 2.7.15 has been released. The update includes several new features like Multiple Alignment View, Active Objects Inspector, and binary packages for Linux OpenSUSE 13.1. The release notes include more information on features, fixes and improvements.
New CDD Release v.3.11 includes recomputed PSSMs and more
Wednesday, February 19, 2014
Conserved Domain Database (CDD) version 3.11 is now available with 596 new or updated NCBI-curated and 49,641 total domain models. The new version now contains the most recent Pfam release 27.
Updates to the Conserved Domain Database include:
- Position-specific score matrices (PSSMs) have been recomputed for many models in CDD, and frequency tables have been added to the PSSMs;
- The search databases distributed as part of this release can now be used with the more recent versions of RPS-BLAST (BLAST release 2.2.28 and up) using composition-based scoring. This abolishes the need to mask out compositionally biased regions in query sequences;
- Domain annotation displays in CD-Search, BATCH CD-Search, and other services now all use a uniform display style. A new display option in CD-Search and BATCH CD-Search provides “standard” results, in addition to “concise” and “full” results. “Standard” results will provide, for each region on the query sequence, the best0-scoring domain model (if any) from each of CDD’s database providers (Pfam, SMART, COG, TIGRFAMs, Protein Clusters, and the NCBI in-house curation project), but will suppress redundancy from within a single provider's results list.
You can access CDD at the Conserved Domains homepage and find updated content on the CDD FTP site.
GenBank has milestone 200th release
Tuesday, February 18, 2014
GenBank's 200th release is now available through NCBI’s Entrez and BLAST services.
Release 200.0 (2/12/2014) has 171,123,749 non-WGS, non-CON records containing 157,943,793,171 base pairs of sequence data. In addition, there are 139,725,795 WGS records containing 591,378,698,544 base pairs of sequence data.
During the 64 days between the close dates for GenBank Releases 199.0 and 200.0, the non-WGS/non-CON portion of GenBank grew by 1,713,261,609 base pairs and by 1,792,342 sequence records. During that same period, 4,979,722 records were updated. An average of 105,813 non-WGS/non-CON records were added and/or updated per day. Between releases 199.9 and 200.0, the WGS component of GenBank grew by 34,614,377,046 base pairs and by 5,907,225 sequence records.
The total number of sequence data files increased by 34 with this release. The divisions are as follows:
- BCT: 4 new files, now a total of 118 files
- CON: 11 new files, now a total of 242 files
- GSS: 5 new files, now a total of 284 files
- INV: 2 new files, now a total of 38 files
- PAT: 5 new files, now a total of 204 files
- PLN: 2 new files, now a total of 67 files
- PRI 1 new file, now a total of 47 files
- TSA 3 new files, now a total of 150 files
- VRT: 1 new file, now a total of 32 files.
For downloading purposes, please keep in mind that the GenBank flatfiles are approximately 625 GB (sequence files only). The ASN.1 data are approximately 522 GB.
More information about GenBank Release 200.0 and coming changes are available in the release notes.
Human genome annotation release 106 now available
Tuesday, February 11, 2014
The human (Homo sapiens) genome annotation has recently been updated to annotation release 106. The data is now available in the Nucleotide, Protein and Gene databases, is searchable using BLAST, and can be downloaded from the FTP site.
The annotated assemblies include GRCh38 (GCF_000001405.26), which was published in December 2013 and represents a major, chromosome-coordinate changing update to the reference assembly (for more information, visit the GRC). Annotation release 106 also includes a re-annotation of the assemblies CHM1_1.1 (GCF_000306695.2) and HuRef (GCF_000002125.1).
Some highlights of the GRCh38 annotation results (as compared to annotation release 105 of the previous assembly, GRCh37.p13) include:
- A total of 29,399 genes are predicted (an increase of 5.6%)
- A total of 69,826 protein-coding transcripts are annotated (an increase of 3.4%)
- The number of CDSs annotated as partial decreased from 96 to 56
- The number of curated RefSeq transcripts with an alignment split across scaffolds decreased from 30 to 5
- The number of protein coding genes found only on alternate loci and/or novel patches increased from 28 to 64.
There are significant improvements to gene annotation on the new GRCh38 assembly. For example:
- SRGAP2: Previously split across scaffolds with an inversion
- DPP6: Previously split across scaffolds
- EPPK1: Previously internally partial
- DOC2B: Previously 3’ partial
See what other annotation runs are in progress on the Eukaryotic Genome Annotation Pipeline status page.
NCBI releases Entrez Direct, the Entrez utilities on the UNIX command line
Thursday, February 06, 2014
NCBI has just released Entrez Direct, a new software suite that enables users to use the UNIX command line to directly access NCBI databases, as well as to parse and format the data to create customized downloads.
For over a decade, researchers have been able to access Entrez search, retrieval and linking functions through the web interface on the NCBI site or through Entrez Utilities (E-Utilities), the public API for Entrez. Entrez Direct now brings Entrez to the UNIX command line by offering a set of UNIX executables that call the E-utilities directly and provide a variety of post-processing functions. Using these functions, arbitrary fields from complex record formats can be parsed and converted to simple tables suitable for importing into spreadsheets or external databases.
Entrez Direct is available as a simple FTP download and has extensive documentation on the NCBI web site. The package itself consists of a master Perl script and several UNIX shell scripts that serve as an interface to the Perl script. Therefore, Entrez Direct will run on UNIX and Macintosh computers that have the Perl language installed, and under the Cygwin UNIX-emulation environment on Windows PCs.
The Entrez Direct documentation provides several examples of how these programs may be used, and we have provided three additional examples below. Links are provided to the commands and output files.
1. Retrieve a set of PubMed abstracts
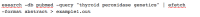
Figure
This command searches PubMed with the query “thyroid peroxidase genetics” and then downloads the retrieved records as abstracts, which are then written to a local file.
2. Download a table of start and stop coordinates for all genes on human chromosome 17 from all available assemblies.

Figure
This command demonstrates the parsing function “xtract” that is unique to Entrez Direct.
First, “esearch” retrieves all current genes on human chromosome 17, and these results are passed to “esummary”. The resulting XML document summaries are passed to “xtract”, which parses the assembly and gene coordinate details and writes them to an output file. The first few lines of this file are shown below.

Figure
This file contains data segmented for each gene and begins with a line containing the Gene ID, followed by lines with these columns: annotation release, assembly accession.version, chromosome accession.version, start coordinate, stop coordinate.
3. Download a table of start and stop coordinates for all genes on human chromosome 17 from the current reference assembly.
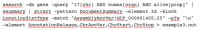
Figure
This command is an extension of Example 2 in that it uses the “-match” option to limit the output to data from the current reference assembly (GCF_000001405.25).

Figure
The output file has the same format as that for Example 2, except that column 2 (assembly accession.version) is omitted.
We will be posting additional Entrez Direct examples and explanations on the NCBI Insights blog and in the Entrez Direct documentation, and will continue to announce updates on the utilities-announce list.
For more information:
Sequence Viewer updated to version 3.1
Tuesday, February 04, 2014
NCBI Sequence Viewer provides a graphical view of sequences and color-coded annotations on regions of sequence stored in the Nucleotide and Protein databases. Sequence Viewer has recently been updated and now has improved compatibility with Internet Explorer for users embedding Sequence Viewer on their web sites and improved performance and response time.
A full list of new features, improvements and fixes is included in the release notes.
- NCBI News, February 2014 - NCBI NewsNCBI News, February 2014 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...
