NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
BLAST machine image (AMI) hosted at Amazon Web Services
Thursday, June 26, 2014
The NCBI now has a BLAST installation at Amazon Web Services, as part of an effort to deliver services to users with new cloud technologies. The installation can be accessed as an Amazon Machine Image (AMI), which allows users to run stand-alone searches with the BLAST+ applications, submit searches through a subset of the NCBI-BLAST URL API, and perform searches with a simplified web page. The AMI also includes a FUSE client that can download BLAST databases during the first search.
For information about the AMI and links to documentation, please see this page.
Green monkey annotation release 100 now available
Monday, June 23, 2014
The green monkey (Chlorocebus sabaeus) annotation is now accessible in the Nucleotide, Protein sequence and Gene databases, searchable using BLAST, and downloadable from the FTP site.
Chlorocebus sabaeus annotation release 100, based on the sequence assembly Chlorocebus_sabaeus 1.1 (GCF_000409795.2) identifies a total of 29,648 genes. This annotation used 22 billion short reads, from 179 distinct BioSample accessions, available from the Sequence Read Archive to assist in gene prediction. This large amount of short reads allowed the identification of alternative variants for over 13,000 genes.
More statistics are available in the Chlorocebus sabaeus Annotation Release 100 Report.
See what other annotation runs are in progress on the Eukaryotic Genome Annotation Pipeline status page.
NCBI's latest YouTube video explores Variation Viewer
Wednesday, June 18, 2014
The most recent video from NCBI demonstrates the basic functions of the Variation Viewer, a tool for navigating variant data in dbSNP, dbVar and ClinVar in a genomic context.
GenBank release 202.0 is now available via FTP
Tuesday, June 17, 2014
Release 202.0 (6/12/2014) has 173,353,076 non-WGS, non-CON records containing 161,822,845,643 base pairs of sequence data. In addition, there are 175,779,064 WGS records containing 719,581,958,743 base pairs of sequence data.
During the 60 days between the close dates for GenBank Releases 201.0 and 202.0, the non-WGS/non-CON portion of GenBank grew by 2,009,433,883 base pairs and by 1,608,590 sequence records. During that same period, 564,904 records were updated; an average of 36,225 non-WGS/non-CON records were added and/or updated per day. Between releases 201.0 and 202.0, the WGS component of GenBank grew by 98,566,526,306 base pairs and by 32,332,274 sequence records.
The total number of sequence data files increased by 43 with this release. The divisions are as follows:
- BCT: 3 new files, now a total of 136
- CON: 21 new files, now a total of 267
- ENV: 4 new files, now a total of 73
- EST: 1 less file, now a total of 475
- GSS: 2 new files, now a total of 287
- INV: 1 new file, now a total of 39
- PAT: 5 new files, now a total of 209
- PLN: 2 new files, now a total of 70
- TSA: 6 new files, now a total of 156
Note that the loss of one EST data file is not due to removal of EST sequences.
For downloading purposes, please keep in mind that the GenBank flatfiles are approximately 42 GB (sequence files only). The ASN.1 data are approximately 538 GB.
More information about GenBank Release 202.0 and coming changes are available in the release notes.
RefSeq model sequences can now be constructed from genomic and transcript sequences
Friday, June 13, 2014
Software version 6.0 of the Eukaryotic Genome Annotation Pipeline has recently been released. Starting with this release, RefSeq transcript and protein models, which have traditionally been constructed based on the genomic sequence alone, can now be constructed from a combination of the genomic sequence (upon which the model is called) and transcript sequence that compensates for small gaps in the genomic sequence.
This offers a significant improvement in completeness and quality for RefSeq model transcripts and proteins. Sequence records for such models contain the RefSeq attribute “assembly gap” and can be queried in Entrez with “assembly_gap[properties]”.
For example, over 3,900 models benefited from this improvement in the recent green anole annotation release 101. One model transcript, XM_003230654.2, contains three exons which are derived from the component genomic contig AAWZ02039332.1 of scaffold NW_003342544.1 and is also extended at the 5-prime end beyond the end of the scaffold (e.g., into an assembly gap) based on the alignment of transcript GAFK01002911.1. As a result, the transcript XM_003230654.2 and protein XP_003230702.2 are now complete, while the previous versions, XM_003230654.1 and XP_003230702.1, annotated in 2011, were partial.
A detailed description of the sequence records for gap-filled models is in the Eukaryotic Genome Annotation Pipeline software release notes.
More information on the RefSeq project can be found here, and more information on annotation runs in progress can be found on the Eukaryotic Genome Annotation Pipeline status page.
Genome Workbench 2.7.19 released
Tuesday, June 10, 2014
Genome Workbench 2.7.19 has been released. The update has several new features, including improved searching and case sensitivity in Text View. The release notes include more information on features, fixes and improvements.
dbSNP human Build 141 now available
Wednesday, June 04, 2014
dbSNP human Build 141, based on the GRCh38 and GRCh37.p13 assemblies, is now available on the integrated NCBI Entrez system and through FTP. Build 141 provides more than 260 million submitted SNP (ss) and over 62 million Reference SNP (rs) clusters. To see complete build statistics, visit the dbSNP summary page. For more information on Build 141, including notes on downloading, policy revisions, and reporting on RefSNP, see this dbSNP listserv announcement.
Update: In addition to the primary assembly unit of assembled chromosones, the dbSNP annotation of GRCh38 and other Genome Reference Consortium (GRC) assemblies will also contain:
- Patch sequences: sequences provided as assembly updates outside of the normal release cycle;
- Alternate loci: sequences that provide an alternative representation of a locus found in a largely haploid assembly;
- PAR sequences: pseudo-autosomal region found on the X and Y chromosomes of mammals; and
- Unplaced sequences: sequences found in an assembly that are not associated with any chromosome.
For more detailed definitions of these sequences, visit the GRC Assembly Terminology page.
Although interesting genetic variations may occur in these sequences, most researchers may never see them, due to current VCF file specification limits. VCF files are a file format used by the 1000 Genomes Project to store genomic variation information. To support reporting on these additional, non-primary chromosome locations, NCBI has released companion files for clinical data, named with a "papu" (patch, alternate, PAR, unplaced) extension. The files are available on the dbSNP FTP sites for GRCh38 and GRCh37.p13. Please note that GRCh38 currently does not have PATCH sequences. VCF files will be updated when these sequences are released for GRCh38.
New features simplify access to annotation information in NCBI's Gene
Tuesday, June 03, 2014
NCBI’s Gene resource is pleased to announce several new features aimed at providing easier access to annotation information.
First, the “Genomic context” section for genes annotated using NCBI's Eukaryotic Genome Annotation Pipeline, including human, mouse, and 130 other species, has been restructured to include a table that includes the Annotation Release, Assembly, and sequence Location. The table provides a convenient view of the location on the reference primary assembly.
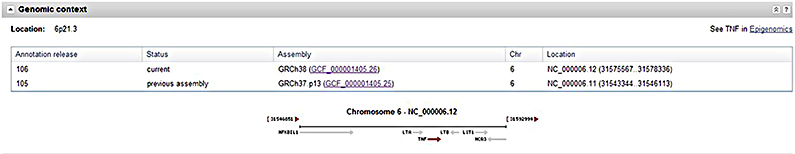
Second, to make it easier for users working with previous assembly versions, the same table mentioned above includes the sequence location from the last annotation of the previous assembly version. This feature is currently limited to human, where it displays the location on the GRCh37.p13 assembly. It will be expanded to more organisms with future assembly updates.
Third, the "Genomic regions, transcripts, and products" section also includes the last assembly in the pulldown menu, giving you access to the prior annotation in the graphical view and through the "Go to nucleotide:" links.
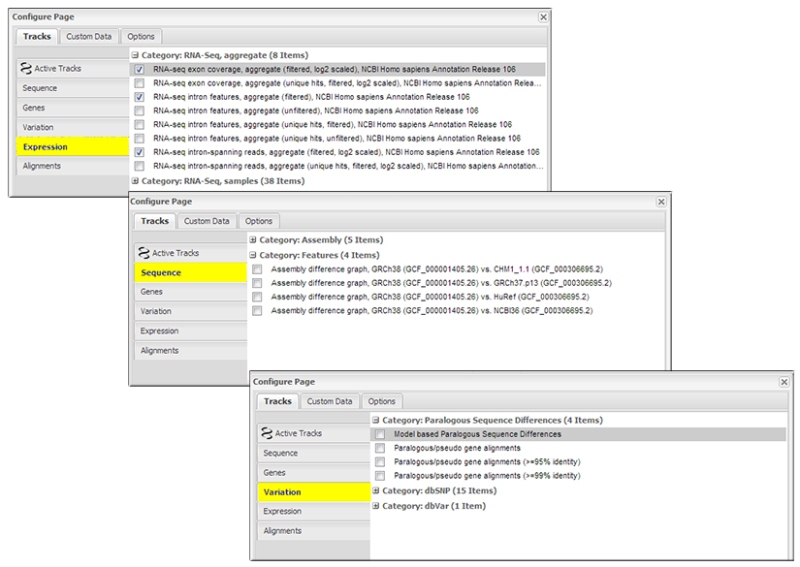
Fourth, one of the most exciting changes can be found in the Graphical View "Configure" page, accessed using the button in the upper right corner of the graphic. This interface now provides access to many more tracks than were previously available, including:
- Under the "Expression" tab, RNA-seq expression tracks computed for each individual BioSample that were aligned as part of the annotation process. These data can provide valuable information about differential expression in tissues or developmental stages. RNA-seq tracks are currently available for 65 taxa that have been annotated using NCBI's Eukaryotic Genome Annotation Pipeline.
- Under the "Sequence" tab, Assembly difference graphs that highlight differences between the GRCh38 and other human assemblies.
- Under the "Variation" tab, paralogous gene alignment tracks that show the alignment of paralogous gene features. These tracks are a useful view of how similar a gene is to any pseudogenes or other paralogs that are annotated in the genome.
Fifth, the Variation section for human genes includes links to the new Variation Viewer genome browser, using either the GRCh37.p13 or GRCh38 assemblies.
Please stay tuned as documentation is updated to reflect these new changes.
- BLAST machine image (AMI) hosted at Amazon Web Services
- Green monkey annotation release 100 now available
- NCBI's latest YouTube video explores Variation Viewer
- GenBank release 202.0 is now available via FTP
- RefSeq model sequences can now be constructed from genomic and transcript sequences
- Genome Workbench 2.7.19 released
- dbSNP human Build 141 now available
- New features simplify access to annotation information in NCBI's Gene
- NCBI News, June 2014 - NCBI NewsNCBI News, June 2014 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...
