NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
General Research Use collection streamlines access to patient-level data in dbGaP
Tuesday, July 29, 2014
In response to many requests from dbGaP users to simplify and streamline the data access request process while respecting patient consent, dbGaP staff have identified “General Research Use” individuals from different studies and created a collection that allows users to access data on these individuals through a single access request.
Most studies in dbGaP have a significant fraction of participants who consented for “General Research Use." NIH recognizes that the consents for these study participants are essentially the same, even though the individuals participated in different studies. Therefore, NIH decided to create a streamlined process that would allow users to obtain data on the collection of the individuals who consented for “General Research Use” in one single request.
Investigators approved for access to the datasets within the Collection with have access to the data for the standard one-year approval period. While you may only wish to use data on some of the individuals for your research, approved users will have access to data on the full set of individuals. In addition, any new individuals in the “General Research Use” category will be automatically added to this collection; you will not need to make another request to make use of the data relating to these new individuals during your one-year approval period.
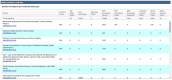
Figure
Figure 1. A sample of the studies included in the dbGaP General Research Use collection.
You can obtain access to this collection (currently 71 studies) through a single access request for "dbGaP Collection: Compilation of Individual-Level Genomic Data for General Research Use." The datasets included in this collection have been designated as appropriate for general research use (GRU) by submitting institutions, which indicates that there are no further limitations on secondary research use beyond those outlined in the Genomic Data User Code of Conduct.
To help expedite the processing of requests for access to the Collection, the NIH review will be conducted by a central Data Access Committee. GRU-designated datasets will be added to the study only after the publication embargo for the original study has expired.
For more information, visit the dbGaP study page.
NCBI/CDC/FDA/USDA collaboration using whole genome sequencing (WGS) to improve food safety is honored with an HHSinnovates award
Tuesday, July 22, 2014
A collaborative project between NCBI and several other Federal and state partners to reduce the time and improve the accuracy of detecting foodborne pathogens by using whole genome sequencing (WGS) techniques received an HHSinnovates award on July 21.
The HHSinnovates program was initiated in 2010 to recognize new ideas and solutions developed by HHS employees and their collaborators. Six finalist teams were recognized at the awards ceremony. The WGS Food Safety Project, which also involved the Centers for Disease Control and Prevention (CDC), the Food and Drug Administration (FDA), the U.S. Department of Agriculture (USDA), and state public health laboratories, was one of three projects to be honored as “Secretary's Picks” by HHS Secretary Sylvia Mathews Burwell.
Presenting the award, HHS Deputy Secretary Bill Corr said: “Together all these folks engaged in a demonstration project to showcase the benefits of using whole genome sequencing for food surveillance and detection purposes. They showed that whole genome sequencing can produce faster detection of foodborne pathogens than the traditional method, helping us stop an outbreak of disease in its tracks, and for that we deeply appreciate your work.” The award went to the specific individuals leading the project in the various agencies; in the case of NCBI, Senior Scientist William Klimke, Ph.D., was honored for his work in heading NCBI’s part of the project.
WGS provides greater specificity than other techniques, such as the commonly used pulsed-field gel electrophoresis (PFGE), in identifying the DNA fingerprint of bacteria. It also can more rapidly determine whether isolates are related to a foodborne disease outbreak.
The demonstration project involves real-time sequencing of Listeria monocytogenes isolates from human DNA as well as the food supply chain. In the project, the whole genomes of isolates are sequenced and the sequencing data are sent to NCBI, which performs assembly, annotation, and analysis and then sends results back to CDC, FDA, USDA, and the labs.
Collaborative projects using WGS for other pathogens related to food safety are also underway.
Major revision of the NCBI genomes FTP site this summer
Friday, July 18, 2014
Within the next two weeks, NCBI will make a major revision to the genomes FTP site. This redesign will expand available content and facilitate data access through an organized, predictable directory hierarchy. The updated site will also provide greater support for downloading assembled genome sequences and/or corresponding annotation data. To give those with automated tools time to update, we plan to maintain the older content and structure of the preexisting /genomes/ FTP site in parallel with the new structure for six months.
The new FTP site structure provides a single entry point to access content representing either GenBank or RefSeq data. Advantages of the updated genomes FTP site include the comprehensive provision of data through a single process flow that is reliant on content in NCBI’s Assembly database (which excludes viruses at this time), integration of quality assurance regression tests, and provision of a consistent core set of files for all organisms and assemblies.
Stay tuned to the NCBI News site and our social media accounts (Facebook, Twitter, LinkedIn, NCBI Announce listserv) for more information about the changes to come and the official launch of the revamped genomes FTP site.
NCBI webinar "Using the New NCBI Variation Viewer to Explore Human Genetic Variation" on August 13th
Wednesday, July 16, 2014
On August 13th, NCBI will host a webinar entitled “Using the New NCBI Variation Viewer to Explore Human Genetic Variation”. This presentation will show you how to find human sequence variants by chromosome position, gene, disease names and database identifiers (RefSNP, Variant region IDs) using NCBI’s new Variation Viewer.
You will learn how to browse the genome, navigate by gene or exon, filter results by one or more categories including allele frequencies from 1000 Genomes or GO-ESP, and link to related information in NCBI’s molecular databases and medical genetic resources such as ClinVar, MedGen, and GTR. You will also be shown how to upload your own data to add to the display and download results.
Anyone who works with clinical or research variation data will find that the Variation Viewer provides a convenient and powerful way to access human variation data in a genomic context that is fully integrated with all other NCBI tools and databases.
To register, please go to this link: https://attendee.gotowebinar.com/register/2762824590748330498.
RefSeq release 66 available on FTP site
Thursday, July 10, 2014
The full RefSeq release 66 is now available with nearly 59 million records describing 43,671,159 proteins, 7,568,770 RNAs, and sequences from 41,263 different NCBI Taxons.
More details about the RefSeq release 66 are included in the release statistics and release notes. In addition, reports indicating the accessions included in the release and the files installed are available.
NCBI's latest YouTube video presents special features in SciENcv
Monday, July 07, 2014
NCBI’s latest YouTube video focuses on special features in SciENcv (Science Experts Network Curriculum Vitae) that help users create, share, and maintain NIH Biosketch profiles for federal grant applications.
While this video centers on a specific case, anyone with a My NCBI account may use SciENCv. For more general information on SciENcv, click on these links:
"BLAST in the Cloud!" webinar on July 30th showcases NCBI-BLAST Amazon Machine Image
Tuesday, July 01, 2014
As stated on June 26, web and standalone BLAST are now available on Amazon Web Services (AWS). On July 30, 2014, NCBI will offer a webinar entitled “BLAST in the Cloud”. This presentation will show you how to log on to AWS and deploy the NCBI-BLAST Amazon Machine Image (AMI) quickly. The BLAST AMI includes the BLAST+ applications, a client that can download databases from the NCBI, a web application that implements a subset of the NCBI URL API, and a simplified BLAST search webpage. Prior knowledge of using web and standalone BLAST is required.
To register, please go to this link: https://attendee.gotowebinar.com/register/8126572163773355778.
- General Research Use collection streamlines access to patient-level data in dbGaP
- NCBI/CDC/FDA/USDA collaboration using whole genome sequencing (WGS) to improve food safety is honored with an HHSinnovates award
- Major revision of the NCBI genomes FTP site this summer
- NCBI webinar "Using the New NCBI Variation Viewer to Explore Human Genetic Variation" on August 13th
- RefSeq release 66 available on FTP site
- NCBI's latest YouTube video presents special features in SciENcv
- "BLAST in the Cloud!" webinar on July 30th showcases NCBI-BLAST Amazon Machine Image
- NCBI News, July 2014 - NCBI NewsNCBI News, July 2014 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...
