NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
NCBI Sequence Viewer version 3.4 available
Tuesday, September 30, 2014
NCBI Sequence Viewer has recently been updated and now has improved visualization of graphs, sequence track and other text, as well as a reworked configuration dialog. A full list of new features, improvements and fixes is included in the release notes.
Sequence Viewer provides a graphical view of sequences and color-coded annotations on regions of sequences stored in the Nucleotide and Protein databases.
HIV-1, human interaction database updated
Monday, September 29, 2014
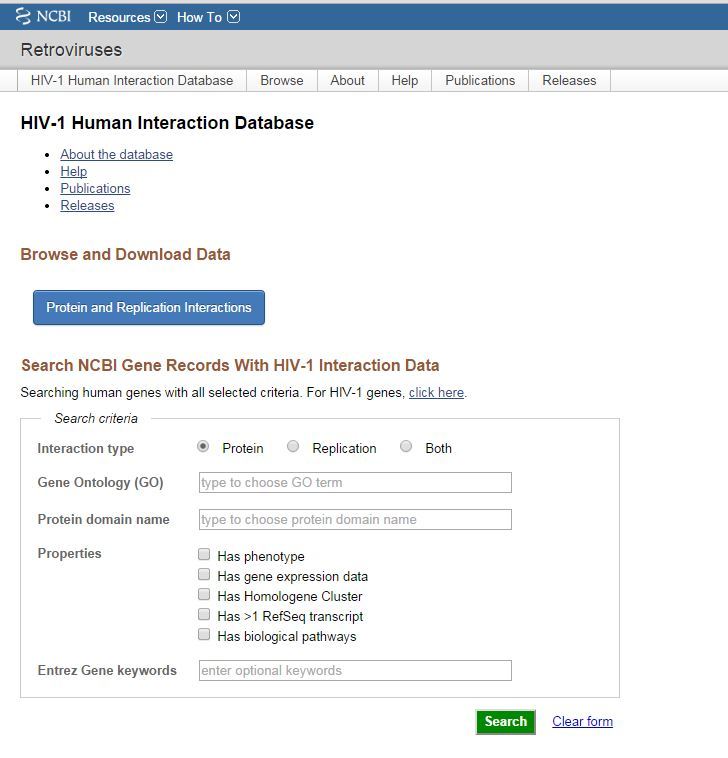
The HIV-1, human interaction database has been updated and is now on an improved page. The improved interface includes help documentation and supports structured queries against Gene, as well as browsing, filtering and downloading the protein and replication interaction data sets. The most recent data release (June 2014) includes 12,785 HIV-1, human protein-protein interactions for 3,142 human genes and 1,316 replication interactions for 1,250 human genes.
Figure 1. The HIV-1 interactions database homepage.
The HIV-1, human interactions project collates published reports of two types of interactions: HIV-1, human protein interactions, and human gene knock-downs that affect virus replication which are reported as “replication interactions”.
Virus Variation Resource pages for Ebolavirus, MERS coronoavirus give quick and easy access to related sequences and other data
Friday, September 19, 2014
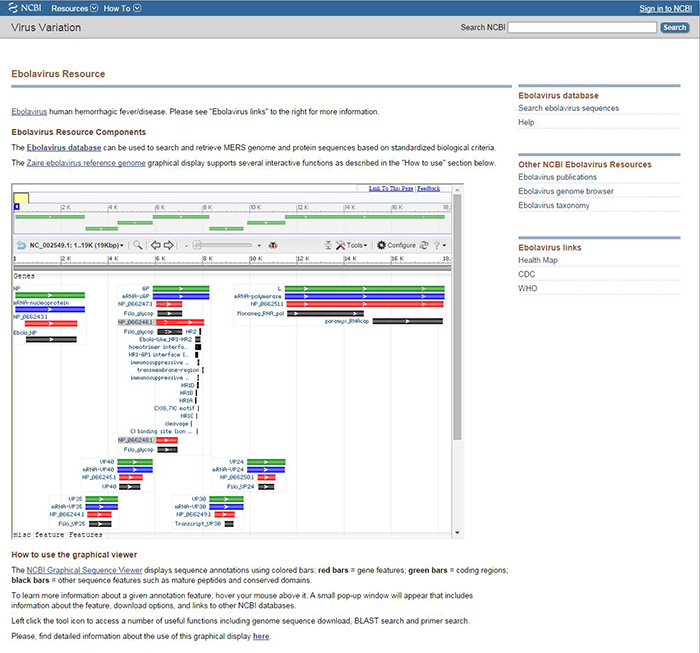
NCBI has created resource pages for Ebolavirus and MERS coronavirus, giving users an easy way to find all sequences related to these pathogens. These pages aggregate links to virus data at NCBI and also provide important links out to other information at the CDC, WHO, and HealthMap.
Figure 1. Ebolavirus Resource page. The main column gives a brief description of the resource and displays the NCBI Graphical Sequence Viewer. In the right column, from top to bottom, are links to: the Ebolavirus database, other NCBI Ebolavirus resources, and links out to HealthMap, CDC, and WHO. The MERS coronavirus resource page has the same layout and links.
The Virus Variation resource pages all include a description of the resource components: the database, the reference genome graphical display and links to other resources, both external and within NCBI.
Figure
Figure 2. Ebolavirus database. Users can get sequences by accession or browse by searching for a keyword, host, or region/country, among other options.
Dedicated Virus Variation databases for Ebolavirus and MERS coronavirus have been developed. These databases allow searching for nucleotide and protein sequences by a variety of criteria including host, sequence patterns, region or country of isolation, and collection or release dates. The databases allow you to:
- Quickly find the sequences you need, through an intuitive search interface for all viral sequences using standardized protein/gene names and metadata
- Select the latest sequences based on date criteria or sorting of results
- Download sequences in many formats or find links to sequences in NCBI databases.
Visit the Virus Variation homepage to see resource pages for other viruses.
Simplified FASTA headers included on new NCBI Genomes FTP site
Wednesday, September 17, 2014
Last month, a major revision of the NCBI Genomes FTP site was announced. In response to user feedback, a new format for FASTA headers of genome, protein and transcript records has been implemented. This new format is limited to records in the /all/, /refseq/, and /genbank/ directories on the new Genomes FTP site and does not affect the Nucleotide database web FASTA displays.
Now, instead of ">gi|xx|dbsrc|accession.version|description", the new format is simply ">accession.version description".
For example, the header on the FASTA record for Homo sapiens chromosome 1 was previously:
>gi|568336023|gb|CM000663.2| Homo sapiens chromosome 1, GRCh38 reference primary assembly.
On the new Genomes FTP site, the header is now:
>CM000663.2 Homo sapiens chromosome 1, GRCh38 reference primary assembly.
NCBI has traditionally used a compound FASTA sequence identifier string in which multiple IDs were separated by “|” characters. This format provides more information, but requires that the individual sequence identifiers be parsed out of the compound string. The simpler sequence identifier string is identical to that used in the GFF annotation files on the genomes FTP site. Providing sequence and annotation files with matching sequence identifiers supports their use in commonly used RNA-Seq analysis packages and in other analysis pipelines that rely on simple string comparison to match sequence identifiers.
More information about the revised Genomes FTP site, including the new FASTA header format, is available on the Genomes Download FAQ page.
RefSeq release 67 available on FTP
Thursday, September 11, 2014
The full RefSeq release 67 is now available on the FTP site with over 61 million records describing 45,166,402 proteins, 8,163,775 RNAs, and sequences from 41,913 different NCBI TaxIDs.
More details about the RefSeq release 67 are included in the release statistics and release notes. In addition, reports indicating the accessions included in the release and the files installed are available.
Identical Protein Report Display option added to Protein database
Tuesday, September 09, 2014
A new display option has been added to the Protein database - the "Identical Protein Report". When viewing an individual record, this display allows you to access a list of all other identical proteins including those submitted as translations to GenBank, as well as RefSeq, UniProtKB/Swiss-Prot, PIR, PDB, and patented protein records.
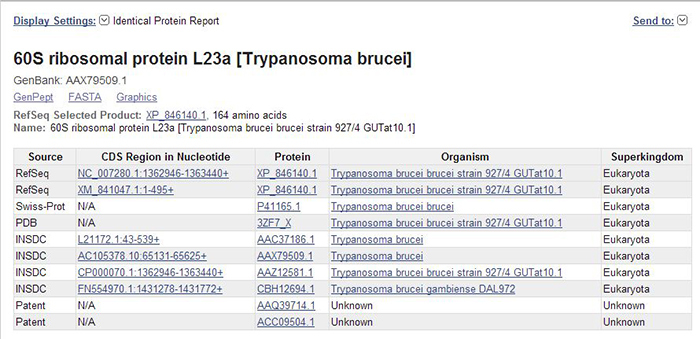
Figure 1. The “Identical Protein Report” display setting in the Protein database, showing identical proteins for 60S ribosomal protein L23 [Trypanosoma brucei].
As shown in Figure 1, the page title reflects the protein record from which you started. Beneath that, there is information on the suggested RefSeq preferred protein accession, protein length, and protein name. Identical proteins are presented in a tabular format that includes information on the database source (e.g., RefSeq, INSDC, etc.), the corresponding nucleotide CDS accession and location, the organism name, and the superkingdom. The displayed table can be downloaded for further use, and is also available through Eutils.
The Identical Protein Report display setting provides important functions, such as:
- A mapping table between protein accessions and the nucleotide record(s) on which they are annotated, when relevant;
- For the RefSeq autonomous non-redundant protein dataset, a mapping table to the organisms to which the protein is relevant;
- And identification of highly conserved proteins when the identical protein sequence is found annotated on divergent species.
- NCBI Sequence Viewer version 3.4 available
- HIV-1, human interaction database updated
- Virus Variation Resource pages for Ebolavirus, MERS coronoavirus give quick and easy access to related sequences and other data
- Simplified FASTA headers included on new NCBI Genomes FTP site
- RefSeq release 67 available on FTP
- Identical Protein Report Display option added to Protein database
- NCBI News, September 2014 - NCBI NewsNCBI News, September 2014 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...