NCBI to hold two-day genomics hackathon in January
Wednesday, November 26, 2014
From January 5th to 7th, NCBI will host a genomics hackathon focusing on advanced bioinformatics analysis of next generation sequencing data. This event is for students, postdocs and investigators already engaged in the use of pipelines for genomic analyses from next generation sequencing data. Working groups of 5-6 individuals will be formed for DNA-Seq/multiomics, RNA-seq, metagenomics and Epigenomics. These groups will build pipelines to analyze large datasets within a cloud infrastructure.
Organization
After a basic organizational session, teams will spend 2.5 days analyzing a challenging set of scientific problems related to a group of datasets. Students will analyze and combine datasets in order to work on these problems. This course will take place on the NIH main campus in Bethesda, Maryland.
Datasets
Datasets will come from the public repositories housed at NCBI. During the course, students will have an opportunity to include other datasets and tools for analysis. Please note, if you use your own data during the course, we ask that you submit it to a public database within six months of the end of the event.
Products
All pipelines and other scripts, software and programs generated in this course will be added to a public GitHub repository designed for that purpose. A manuscript outlining the design of the hackathon and descripting participant processes, products and scientific outcomes will be submitted to an appropriate journal.
Application
To apply, complete this form (approximately 10-15 minutes to complete). Applications are due December 1st by 5pm EST. Participants will be selected from a pool of applicants; prior students will be given priority in the event of a tie. Accepted applicants will be notified on December 10th by 9am EST, and have until December 12th at noon to confirm their participation. Please include a monitored email address, in case there are follow-up questions.
Note: Students will need to bring their own laptop to this program. A working knowledge of scripting (e.g., Shell, Python) is necessary to be successful in this event. Employment of higher level scripting or programming languages may also be useful. Also note that the course may extend into the evening hours on Monday and/or Tuesday. Please make any necessary arrangements to accommodate this possibility.
Please contact ben.busby@nih.gov with any questions.
NCBI BioSample includes curated list of over 400 known misidentified and contaminated cell lines
Monday, November 24, 2014
The NCBI BioSample database now includes a curated list of over 400 known misidentified and contaminated cell lines. Scientists should check this list before they start working with a new cell line to see if that cell line is known to be misidentified.
Continuous cell lines are used widely in research as model systems for normal cellular processes and disease states. However, as noted by many (e.g. PubMed 23235867, 20143388, 19003294, 18072586, and 17522957), cell line cross-contamination or misidentification represents a serious and widespread problem, and researchers should take great care to check that their cell line is what they think it is. Cell lines can be easily mislabeled or become overgrown by cells derived from a different individual, tissue or species.
This problem is so common it is thought that thousands of misleading and potentially erroneous papers have been published using cell lines that are incorrectly identified (PubMed 20448633). The first step in combating this problem is to make sure your cell line is not on the list of known misidentified and cross-contaminated cell lines. Detailed information about how to test your cell lines is provided by the International Cell Line Authentication Committee.
NCBI Eukaryotic Genome Annotation Pipeline breaks record; over 100 organisms annotated this year
Thursday, November 20, 2014
The NCBI Eukaryotic Genome Annotation Pipeline has broken a record and completed the annotation of over 100 organisms since the beginning of 2014!
As of today, 81 of this year's 104 annotation releases in RefSeq were first annotations, while 23 were updates. RNA-Seq data was used for gene prediction for 73 of the 104 organisms.
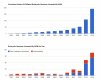
Figure
Figure 1: Top: Cumulative number of different eukaryotic genomes annotated by NCBI. Bottom: Eukaryotic genomes annotated by NCBI per year.
Related links:
- Request a genome annotation
- Browse all eukaryotes annotated by NCBI
NCBI BankIt webinar on December 17th
Thursday, November 20, 2014
On December 17th, NCBI will have a webinar entitled "A Submitter's Guide to GenBank: Using BankIt for Small-Scale Nucleotide Sequence Submissions". This presentation will outline the process of using BankIt, a web-based submission tool at NCBI, to submit sequence data to the GenBank database.
Presenters will demonstrate how to use BankIt forms to complete a submission of a single or a few nucleotide sequences, and how to format and upload text input files needed for submissions of multiple sequences or for sequences with multiple genes.
This webinar will stay at a basic level for sequence submissions; future webinars that illustrate more complex sequence submissions will be considered, depending on the feedback received from this presentation.
To register, visit the webinar registration page. To see materials and videos from previous webinars, as well as descriptions of upcoming webinars, see the NCBI Webinars page.
NCBI E-Utilities webinar video now on YouTube
Thursday, November 13, 2014
October's webinar, "An Introduction to NCBI's E-Utilities, an NCBI API", is now on YouTube and has been added to the NCBI Webinars playlist.
For more information about NCBI's webinars including descriptions of upcoming webinars and materials for past presentations, please see the Webinars homepage.
BLAST URL domain change in effect December 1
Wednesday, November 12, 2014
As announced previously, BLAST searches sent to the www.ncbi.nlm.nih.gov/blast URL will not function as of December 1, 2014. The officially supported URL domain for BLAST searches at the NCBI is blast.ncbi.nlm.nih.gov. Please update your bookmarks, links, and any scripts or applications.
RefSeq release 68 available on FTP
Friday, November 07, 2014
The comprehensive RefSeq release 68 is now available on the FTP site, with over 66 million records describing 46,968,574 proteins, 9,069,704 RNAs, and sequences from 49,312 distinct NCBI TaxIDs.
More details about RefSeq release 68 are included in the release statistics and release notes. In addition, reports indicating the accessions included in the release and the files installed are available.
dbVar releases 1000 Genomes Phase 3 structural variants
Tuesday, November 04, 2014
dbVar has released structural variation (SV) data generated by the 1000 Genomes Project Phase 3 as dbVar study estd214. This large dataset contains SV from 2,500 subjects, and comprises nearly 63,000 variant regions and over 6 million calls, including insertions, deletions, copy number variants (CNVs), mobile element insertions, indels (deletion-insertions), and inversions. The data are available on assemblies GRCh37 (submitted) and GRCh38 (remapped). Genotypes are currently available in VCF.
The data can be accessed from this dbVar Study Page and by FTP.
dbVar releases copy number variation (CNV) data from developmental delay study cited in Nature Reviews Genetics
Monday, November 03, 2014
dbVar recently released copy number variation (CNV) data from a study on dosage-sensitive genes (PMID: 25217958) that was highlighted in Nature Reviews Genetics. In the study, CNV analysis was combined with protein-truncating single-nucleotide variation (SNV) and targeted resequencing to identify dosage-sensitive genes causing developmental delay.
CNV data from Coe et al. (2014) can be accessed at dbVar, and the study itself can be found in PubMed.
Publication Details
Copyright
Publisher
National Center for Biotechnology Information (US), Bethesda (MD)
NLM Citation
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012. NCBI News, November 2014.
