NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
NCBI webinar A Submitter's Guide to GenBank, Part 2 on January 7th
Wednesday, December 31, 2014
On January 7th, NCBI will present the continuation of the December 17th webinar on using BankIt for GenBank submissions. Part 2 will cover how to use BankIt file inputs to complete a submission of a single or a few nucleotide sequences that require multiple features for each sequence. We will also describe how to create and use Feature Table files to add information about sequence data.
This webinar will stay at a basic level for sequence submissions, but future webinars that illustrate more complex sequence submissions will be considered depending on the feedback received from this presentation.
To register, click here. To see materials and videos from previous webinars, as well as descriptions of upcoming webinars, see the NCBI Webinars page.
GenBank release 205.0 is now available via FTP
Tuesday, December 16, 2014
Release 205.0 (12/12/2014) has 179,295,769 non-WGS, non-CON records containing 184,938,063,614 base pairs of sequence data. In addition, there are 200,301,550 WGS records containing 848,977,922,022 base pairs of sequence data.
During the 55 days between the close dates for GenBank releases 204.0 and 205.0, the non-WGS/non-CON portion of GenBank grew by 3,374,386,696 base pairs and by 973,516 sequence records. During that same period, 614,225 records were updated; an average of 28,868 non-WGS/non-CON records were added and/or updated per day. Between releases 204.0 and 205.0, the WGS component of GenBank grew by 43,428,754,314 base pairs and by 4,251,576 sequence records.
The total number of sequence data files increased by 35 with this release. The divisions are as follows:
- BCT: 7 new files, now a total of 159
- CON: 6 new files, now a total of 292
- ENV: 2 new files, now a total of 78
- GSS: 2 new files, now a total of 289
- INV: 8 new files, now a total of 132
- PLN: 6 new files, now a total of 95
- TSA: 3 new files, now a total of 159
- VRL: 1 new file, now a total of 33
For downloading purposes, please keep in mind that the uncompressed GenBank flatfiles are approximately 688 GB (sequence files only). The ASN.1 data require approximately 562 GB.
More information about GenBank release 205.0 is available in the release notes.
Bald eagle and other bird genome sequence and annotation data publicly available at NCBI
Thursday, December 11, 2014
A series of press releases yesterday, including one by Science Publishing, announced the first findings of the Avian Phylogenomics Consortium, who analyzed genome sequence and annotation data for 48 bird genomes representing all of the bird taxonomic orders. All of the sequenced genomes, along with any annotation provided by the submitter, are available in NCBI resources including Assembly, Nucleotide, Protein, the Sequence Read Archive (SRA), and BLAST, or from species-specific GenBank genomes FTP directories. RNA-Seq data for some of the bird species can be found in SRA.
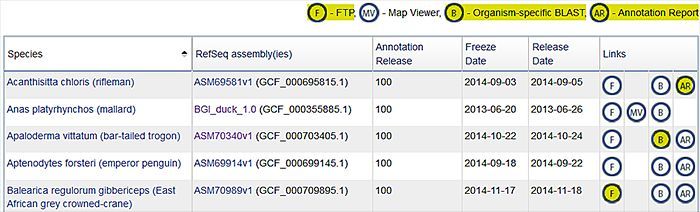
With the exception of three very fragmented assemblies, NCBI annotated the genome assemblies submitted by the Avian Phylogenomics Consortium using NCBI's Eukaryotic Genome Annotation Pipeline, and these annotations are now part of the RefSeq project. The RefSeq project also generated annotations for an additional 6 bird assemblies, for a total of 51 RefSeq genomes. A summary of all the bird genomes that have RefSeq annotation is here.
Figure 1. A selection of the bird genomes with RefSeq annotation. At the top right is a legend describing resource links for each bird genome. Detailed annotation reports, accessible through the "AR" link in the far right column, are available for those genomes annotated in 2014. RefSeq annotation is on organism-specific BLAST pages (the "B" link) and on FTP (the "F" link). Click on the picture to go to the summary table.
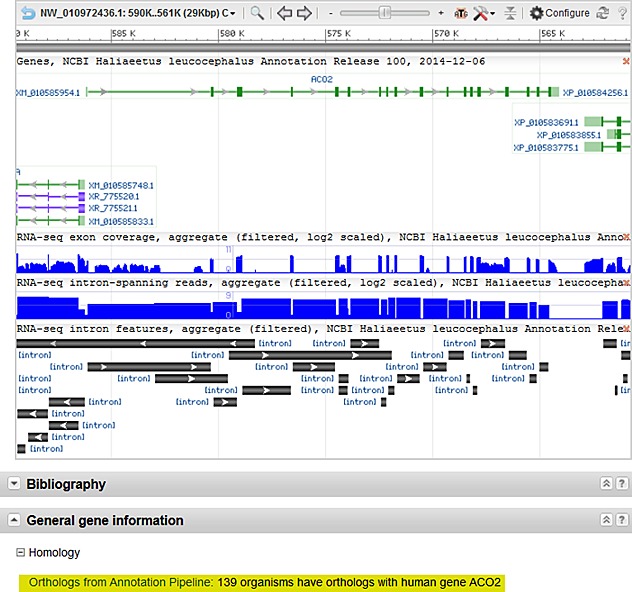
RNA-Seq data was used to generate annotations for 12 of the 51 bird assemblies. The number of protein-coding genes per genome ranges from >13,300 to >21,100 (chicken) with an average of 14,932 protein-coding genes. Orthology to human proteins was also calculated, using simple metrics of local synteny and sequence similarity, and on average, roughly 11,000 orthologous proteins were identified per avian genome. These results are shown in the Homology section of NCBI Gene records (see Figure 2 below).
Figure 2. A portion of the NCBI Gene report for the bald eagle ACO2 gene. The graphical display includes information about the gene structure, the RefSeq transcript and protein models, and RNA-Seq coverage graphs produced by the annotation pipeline. The Homology section is highlighted, showing 139 organisms, including the bald eagle, with orthology to the human ACO2 gene.
Citation Exporter Feature Now Available in PubMed Central
Tuesday, December 09, 2014
PubMed Central (PMC) has added a citation exporter, which makes it easy to retrieve styled citations that you can copy and paste into your manuscripts or download in a format compatible with your bibliographic reference manager software.
When viewing an Entrez search results page, each result summary includes a "Citation" link. When clicked, this will open a pop-up window that you can use to easily copy/paste citations formatted in one of three popular styles: AMA (American Medical Association), MLA (Modern Library Association), or APA (American Psychological Association). In addition, the box has links at the bottom that can be used to download the citation information in one of three machine-readable formats, which most bibliographic reference management software programs can import.
The same citation box can also be invoked from an individual article, either in classic view (with the "Citation" link among the list of formats) or the PubReader view, by clicking on the citation information just below the article title in the banner.
These human-readable styled citations and machine-readable formats will be available through a public API, and we will be providing more details about that in another announcement on the pmc-utils-announce mailing list.
New NCBI Insights blog post: Designing exon-specific primers for the human genome
Tuesday, December 02, 2014
The latest blog post on NCBI Insights shows you how to use NCBI Reference Sequences and Primer-BLAST, NCBI's primer designer and specificity checker, to design a pair of primers that will amplify a single exon, using the human breast cancer 1 gene (BRCA1) as an example.
- NCBI webinar A Submitter's Guide to GenBank, Part 2 on January 7th
- GenBank release 205.0 is now available via FTP
- Bald eagle and other bird genome sequence and annotation data publicly available at NCBI
- Citation Exporter Feature Now Available in PubMed Central
- New NCBI Insights blog post: Designing exon-specific primers for the human genome
- NCBI News, December 2014 - NCBI NewsNCBI News, December 2014 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...