NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
NIHMS's new look streamlines the manuscript submission process
Thursday, January 29, 2015
Today, the NIH Manuscript Submission (NIHMS) system gets a new interface design, as well as updates that streamline the login and manuscript submission processes and provide relevant help information directly on each screen.
Homepage
Figure
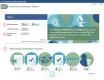
Figure 1. The new NIH Manuscript Submission system homepage.
The NIHMS sign-in routes will now be available from the homepage. Select a route based on your funding agency (1) or sign in through NCBI if you are starting a deposit on an author's behalf(2).
The homepage also includes a graphic overview of the NIHMS process (3). You can hover over each step for more information or click "Learn More" to read the complete overview in the FAQ.
Note: The steps of the NIHMS conversion process will remain the same. An author or PI (i.e., Reviewer) will still need to complete the Initial Approval and Final Approval steps. Updated help documentation and FAQs will help you navigate the process.
Managing Manuscripts
Figure
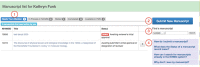
Figure 2. The Manuscript List.
Once you are signed into NIHMS, you will be directed to your Manuscript List. From this page, you can manage and track your existing submissions (1), submit a new manuscript (2), and search for a record (3). You can also click on any headings in the information box (4) to expand a topic and read the help text.
Deposit a Manuscript
The initial deposit still requires you to enter a manuscript and journal title, deposit complete manuscript files, and specify funding information and the embargo.
Figure
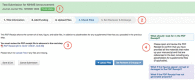
Figure 3. A sample submission.
Key updates include:
- assigning an NIHMSID to a record only after files have been uploaded, i.e., at the Check Files step (1);
- a streamlined deposit process with clearly defined and explained actions in each step (2);
- requiring the Submitter to open the PDF Receipt to review the uploaded files and confirm that the submission is complete before advancing to the next step (3);
- relevant help information available on each page, as in the previous example (4); and
- requiring the Reviewer to add funding before approving the initial deposit (not pictured).
Questions? Contact nihms-helpdesk@ncbi.nlm.nih.gov.
Genome Workbench 2.8.10 available
Monday, January 26, 2015
Genome Workbench 2.8.10 is available, as of January 16th. New features include added support for Ubuntu 14.04 and automatic project save. For the full list of fixes, improvements and features, see the Genome Workbench release notes.
Conserved Domain Database (CDD) version 3.13 now available online and via FTP
Monday, January 26, 2015
Conserved Domain Database (CDD) version 3.13 is now available with 286 new or updated NCBI-curated domains and 50,415 total domain models from CDD’s database providers: Pfam, SMART, COG, TIGRFAMs, Protein Clusters, and the NCBI in-house curation project.
You can access CDD at the Conserved Domains homepage and find updated content on the CDD FTP site. You can also learn about the Conserved Domain Database, how it works and is maintained, as well as future plans for the database in this paper from the most recent Nucleic Acids Research database issue.
NCBI support for SOAP E-Utility ends July 1, 2015
Thursday, January 22, 2015
On July 1, 2015, the NCBI E-Utility SOAP web service, along with the SOAP web service for BLAST, will no longer be supported by NCBI. Any requests to these services after that date will not function.
Please consider using the URL interface to the E-Utilities to retrieve NCBI data or the BLAST REST API to submit BLAST searches.
GenBank surpasses one trillion total bases of publicly available sequence data
Thursday, January 22, 2015
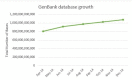
Last October, GenBank (Release 204) exceeded an astounding 1 Terabase of assembled sequence data.
Figure
Figure 1. Total number of bases from April to December 2014.
GenBank is a comprehensive database that contains publicly available nucleotide sequences for over 300,000 formally described species. To learn how NCBI builds GenBank and ensures its uniformity and comprehensiveness, see this recently published paper.
Nucleic Acids Research Database 2015 Issue illustrates NCBI databases, updates and future plans
Wednesday, January 21, 2015
The 22nd annual edition of the Nucleic Acids Research Database Issue features nine papers from NCBI staff that present recent updates to our databases, including GenBank, Gene, and RefSeq.
These papers describe the state of NCBI databases as well as future plans to improve their use, from new reference resources created to improve the usability of viral sequence data to in-house curation efforts in the Conserved Domain Database, and much more.
The NCBI database articles in NAR are also available from PubMed. To read an article, click on the PMID listed below:
- "Database Resources of the National Center for Biotechnology Information" by NCBI Resource Coordinators. (PMID: 25398906)
- "GenBank" by Dennis A. Benson et al. (PMID: 25414350)
- "Gene: a gene-centered information resource at NCBI" by Garth R. Brown et al. (PMID: 25355515)
- "CDD: NCBI's conserved domain database" by Aron Marchler-Bauer et al. (PMID: 25414356)
- "Expanded microbial genome coverage and improved protein family annotation in the COG database" by Michael Y. Galperin, Kira S. Makarova, Yuri I. Wolf and Eugene V. Koonin. (PMID: 25428365)
- "HIV-1, human interaction database: current status and new features" by Danso Ako-Adjei et al. (PMID: 25378338)
- "NCBI Viral Genomes Resource" by J. Rodney Brister, Danso Ako-Adjei, Yiming Bao and Olga Blinkova. (PMID: 25428358)
- "Update on RefSeq microbial genomes resources" by Tatiana Tatusova et al. (PMID: 25510495)
- "Type material in the NCBI Taxonomy Database" by Scott Federhen. (PMID: 25398905)
NCBI YouTube channel: A million views and counting!
Friday, January 16, 2015
As of December 31, 2014, we have passed the 1 million mark for lifetime views on our YouTube channel! The NCBI YouTube channel provides presentations and tutorials about our biomolecular and biomedical literature databases and tools.
Subscribe to our YouTube channel and stay up to date on all the tutorials and webinars we offer.
NCBI's next webinar is The Statistics of Local Pairwise Sequence Alignment, Parts 1 and 2
Tuesday, January 13, 2015
On Thursday, January 22nd, Stephen Altschul of NCBI will present the first part of a discussion of the statistical theory for local sequence alignments like those produced by the BLAST database search programs. It will cover the statistical parameters for local alignment scoring systems, and the formulas for calculating bit scores and asymptotic E-values and p-values from raw alignments scores.
This presentation will continue the following Thursday, January 29th. Part 2 will be a discussion of the considerations that go into the construction and selection of amino acid and nucleic acid scoring systems for pairwise local sequence alignment. It will briefly cover the PAM and BLOSUM series of amino acid substitution matrices, and also the concepts of relative entropy and efficiency for substitution matrices.
E-Utilities users: Keep up to date with changes via the Gene database RSS feed
Monday, January 12, 2015
If you use E-Utilities/ESummary with the Gene database and have not subscribed to the Gene News RSS feed, you probably missed an important announcement about a few impending changes:
A new element in the DocSum has now been added for Gene records. The new Organism element consolidates taxonomic information from the previously used items: Scientific name, common name and TaxID.
The new XML format for ESummary is simpler and more compact, using each field name as the XML tag. For example, <TaxID>9606</TaxID>.
To keep up to date with changes affecting the Gene database, please consider signing up for the Gene Announce RSS feed, which can be found on this page listing all of NCBI's RSS feeds and email listservs.
RefSeq release 69 available on FTP
Wednesday, January 07, 2015
The full RefSeq release 69 is now available on the FTP site with 74 million records describing 52,276,468 proteins, 9,973,568 RNAs, and sequences from 51,661 organisms.
More details about the RefSeq release 69 are included in the release statistics and release notes. In addition, reports indicating the accessions included in the release and the files installed are available.
NCBI annotates 200th eukaryote
Tuesday, January 06, 2015
The NCBI Eukaryotic Genome Annotation Pipeline has passed a new landmark: the completion of the annotation of 200 different organisms including 76 mammals, 51 birds, 26 other vertebrates, 21 invertebrates and 26 plants. Over half of these were annotated with the help of RNA-Seq evidence available in the Sequence Read Archive. The lucky 200th organism is a fish, the large yellow croaker (Larimichthys crocea). See the full list of annotated organisms here, and request the annotation of your favorite!
Data produced by the Eukaryotic Genome Annotation Pipeline is available in the Reference Sequences (RefSeq) collection, BLAST non-redundant and organism-specific databases, Gene database, and on the NCBI FTP site.
NCBI staff will attend International Plant and Animal Genome Conference XXIII
Monday, January 05, 2015
Next week, NCBI staff will present posters and lead a workshop at the International Plant and Animal Genome Conference. In addition, NCBI will have a booth (Booth 618). Staff will be at the booth to answer any questions you may have.
To see a full schedule of NCBI's activities at PAG XXIII, including our annual Genome Resources workshop, click here or visit us at Booth 618.
- NIHMS's new look streamlines the manuscript submission process
- Genome Workbench 2.8.10 available
- Conserved Domain Database (CDD) version 3.13 now available online and via FTP
- NCBI support for SOAP E-Utility ends July 1, 2015
- GenBank surpasses one trillion total bases of publicly available sequence data
- Nucleic Acids Research Database 2015 Issue illustrates NCBI databases, updates and future plans
- NCBI YouTube channel: A million views and counting!
- NCBI's next webinar is The Statistics of Local Pairwise Sequence Alignment, Parts 1 and 2
- E-Utilities users: Keep up to date with changes via the Gene database RSS feed
- RefSeq release 69 available on FTP
- NCBI annotates 200th eukaryote
- NCBI staff will attend International Plant and Animal Genome Conference XXIII
- NCBI News, January 2015 - NCBI NewsNCBI News, January 2015 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...
