NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Variation Viewer 1.4.1 is now available with optimized Variant Filters and Table performance
Thursday, October 29, 2015
Variation Viewer 1.4.1 provides several new features, improvements and bug fixes, including further optimized performance for Variant Filters and Variant Table. A full list of changes to Variation Viewer is available in the release notes.
Variation Viewer is a tool for navigating variant data in dbSNP, dbVar and ClinVar in a genomic context.
New on the NCBI YouTube channel: "LinkOut - Linking to Datasets, Databases and More"
Thursday, October 29, 2015
The recording for the October 2nd webinar "LinkOut - Linking to Datasets, Databases and More" is available on YouTube. The webinar presents an overview of LinkOut and highlights participating resources, with special emphasis on resources beyond full text articles, including databases, datasets and research tools.
Subscribe to the NCBI YouTube channel to receive alerts about new videos ranging from quick tips to full webinar presentations.
OSIRIS Version 2.5 is now available
Wednesday, October 28, 2015
OSIRIS, NCBI's open source short tandem repeat (STR) analysis and quality assurance software package, has just been updated to version 2.5. This version brings several improvements, including multiple peak labels and improved allele and artifact calling. A full list of updates is included in the release notes.
The OSIRIS software for Windows/Mac, the User's Guide, and release notes are all freely available for download on the OSIRIS homepage. In addition, an OSIRIS webinar is available on the NCBI YouTube channel.
OSIRIS was initiated in response to recommendations of a multidisciplinary advisory group empaneled by the U.S. Department of Justice, and was developed in collaboration with state, local and federal forensic laboratories and NIST.
Outdated Genomes FTP directories will be archived on November 30, 2015
Tuesday, October 27, 2015
At the end of November 2015, many outdated Genomes FTP directories will be archived and no longer updated. If you get Genomes data from the NCBI FTP site, please prepare by checking the detailed list of changes below and updating your bookmarks, links and scripts where necessary before November 30, 2015.
FTP directories and files moving on November 30, 2015:
- All directories and files from ftp://ftp.ncbi.nlm.nih.gov/genbank/genomes/ will be archived to ftp://ftp.ncbi.nlm.nih.gov/genomes/archive/old_genbank;
- The following directories from ftp://ftp.ncbi.nlm.nih.gov/genomes/ will be archived to ftp://ftp.ncbi.nlm.nih.gov/genomes/archive/old_refseq/;
- Aedes_aegypti
- Anopheles_gambiae
- Arabidopsis_lyrata
- Arabidopsis_thaliana
- ASSEMBLY_BACTERIA
- Bacteria
- Bacteria_DRAFT
- Branchiostoma_floridae
- Caenorhabditis_elegans
- Chloroplasts
- CLUSTERS
- Drosophila_melanogaster
- Drosophila_pseudoobscura
- Fungi
- Medicago_truncatula
- MITOCHONDRIA
- Physcomitrella_patens
- PLANTS
- Plasmids
- Populus_trichocarpa
- Protozoa
- Sorghum_bicolor
- The file old_genomeID2nucGI from ftp://ftp.ncbi.nlm.nih.gov/genomes/ will be archived to ftp://ftp.ncbi.nlm.nih.gov/genomes/archive;
- The IDS directory from ftp://ftp.ncbi.nlm.nih.gov/genomes/ will be moved to ftp://ftp.ncbi.nlm.nih.gov/genomes/GENOME_REPORTS.
See the NCBI Genomes FTP FAQ for help on using the newer Genomes FTP directories, ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank and ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq that provide replacement content for the directories being archived.
GenBank release 210.0 is now available via FTP
Wednesday, October 21, 2015
GenBank release 210.0 (10/15/15) has 188,372,017 non-WGS, non-CON records containing 202,237,081,559 base pairs of sequence data. In addition, there are 309,198,943 WGS records containing 1,222,635,267,498 base pairs of sequence data, as well as 81,790,031 TSA records containing 70,917,172,944 base pairs of sequence data.
During the 62 days between the close dates for GenBank releases 209.0 and 210.0, the traditional (i.e., non-WGS, non-CON) portion of GenBank grew by 2,413,437,272 base pairs and by 1,306,171 sequence records. During that same period, 280,348 records were updated and an average of 25,573 traditional records were added and/or updated each day.
Between releases 209.0 and 210.0, the WGS component of GenBank grew by 59,359,666,497 base pairs and by 6,243,400 sequence records; the TSA component of GenBank grew by 1,556,418,531 base pairs and by 6,036,982 sequence records.
The total number of sequence data files increased by 38 with this release. The divisions are as follows:
- BCT: 10 new files, now a total of 206
- ENV: 1 new file, now a total of 86
- INV: 3 new files, now a total of 132
- MAM: 9 new files, now a total of 37
- PAT: 6 new files, now a total of 235
- PLN: 5 new files, now a total of 119
- VRL: 1 new file, now a total of 38
- VRT: 3 new files, now a total of 59
For downloading purposes, please keep in mind that the uncompressed GenBank flat files require approximately 742 GB (sequence files only); the ASN.1 data require approximately 605 GB.
More information about GenBank release 210.0 is available in the release notes.
New on the NCBI YouTube channel: Learn how to view track sets and store track collections
Friday, October 16, 2015
Two new videos on the NCBI YouTube channel will show you how to view track sets in all of the NCBI genome browsers and Sequence Viewer displays and how to store and share custom sets of tracks in track collections.
NCBI Recommended Tracks presents track sets, which allow you to instantly tailor your display to a specific need, while My NCBI Track Collections: Introduction shows you how to store and share tracks in custom sets called track collections. If you'd like to learn more about track sets and collections, you can read the FAQ on the Sequence Viewer page.
Subscribe to the NCBI YouTube channel to receive alerts about new videos ranging from quick tips to full webinar presentations.
Larger word size in modified algorithm speeds up BLASTP, BLASTX, TBLASTN search
Thursday, October 15, 2015
The NCBI BLAST webpage now offers faster BLASTP, BLASTX and TBLASTN searches due to a modified algorithm that can use a larger word size. This improvement can make search 2-4 times faster without changing the results most of the time. Please see this article for more details on the modified algorithm.
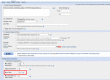
Figure
Figure 1. You can use the word size menu (outlined in red) to recover BLAST search's previous behavior.
Note: You may also recover the search's previous behavior by changing the word size from 6 to 3. To do so, expand "Algorithm parameters" at the bottom of the BLAST page and use the Word size menu (see Figure 1).
Variation Viewer 1.4 is now available with faster filter performance, track sets & collections
Thursday, October 08, 2015
Variation Viewer 1.4 provides several new features, improvements and bug fixes, including significantly faster performance for Sequence Viewer and Variant Table filters, improved documentation readability, added track sets and track collections, and more. A full list of changes to Variation Viewer is available in the release notes.
Variation Viewer is a tool for navigating variant data in dbSNP, dbVar and ClinVar in a genomic context.
Sequence Viewer 3.10 adds support for track sets and track collections, performance optimization and more
Monday, October 05, 2015
Sequence Viewer has been updated to version 3.10, bringing new features, improvements and bug fixes. These changes include:
- Support for track sets and track collections
- Performance optimization allowing faster switching between molecules in genomic browsers and faster Sequence Viewer staging
- A new display option for alignment track
A full list of changes to Sequence Viewer is available in the release notes.
Sequence Viewer is a graphical view of sequences and color-coded annotations on regions of sequences stored in the Nucleotide and Protein databases.
New NCBI Insights blog post: "Troubleshooting GenBank Submissions: Annotating the Coding Region (CDS)"
Friday, October 02, 2015
The latest blog post on NCBI Insights gives GenBank data submitters a workflow for using BLAST to troubleshoot problems with CDS feature annotation. This information is also available in two webinars on the NCBI YouTube channel: Coding Region Annotation and Eukaryotic CDS Annotation.
NCBI Insights is the official NCBI blog, where we share science feature stories, quick tips, and what's new at NCBI.
Subscribe to the NCBI YouTube channel to receive alerts about new videos ranging from quick tips to full webinar presentations.
NCBI staff to attend and present at ASHG 2015
Friday, October 02, 2015
NCBI will participate in the ASHG annual meeting in Baltimore, MD (Oct. 6-10). Staff will participate in the Genome Reference Consortium workshop and present twelve different posters on updated tools and resources for clinical genetics, genomics, and human genome assembly and annotation.
NCBI staff members will also be at the NCBI Exhibit Booth (#2405), where attendees can get answers and provide input for the future development of NCBI human genome resources.
- Variation Viewer 1.4.1 is now available with optimized Variant Filters and Table performance
- New on the NCBI YouTube channel: "LinkOut - Linking to Datasets, Databases and More"
- OSIRIS Version 2.5 is now available
- Outdated Genomes FTP directories will be archived on November 30, 2015
- GenBank release 210.0 is now available via FTP
- New on the NCBI YouTube channel: Learn how to view track sets and store track collections
- Larger word size in modified algorithm speeds up BLASTP, BLASTX, TBLASTN search
- Variation Viewer 1.4 is now available with faster filter performance, track sets & collections
- Sequence Viewer 3.10 adds support for track sets and track collections, performance optimization and more
- New NCBI Insights blog post: "Troubleshooting GenBank Submissions: Annotating the Coding Region (CDS)"
- NCBI staff to attend and present at ASHG 2015
- NCBI News, October 2015 - NCBI NewsNCBI News, October 2015 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...
