NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Genome Workbench 2.10.7 now available
Thursday, June 30, 2016
Genome Workbench 2.10.7 brings a number of new features and fixes like added support for local custom BLAST databases and improvements to Tree View.
For the full list of changes, see the release notes.
July 6th NCBI Minute: Quickly Find Coding Sequences Using ORFfinder
Monday, June 27, 2016
In one week, we’ll show you how to use the redesigned Open Reading Frame Finder (ORFfinder) to quickly identify and analyze complete coding regions on prokaryotic genomic and eukaryotic mRNA sequences.
Date and time: Wednesday, July 6, 2016 12:00 PM EDT
Registration: https://attendee.gotowebinar.com/register/8131176934772138753
After registering, you will receive a confirmation email with information about attending the webinar. After the live presentation, the webinar will be uploaded to the NCBI YouTube channel. Any related materials will be accessible on the Webinars and Courses page; you can also learn about future webinars on this page.
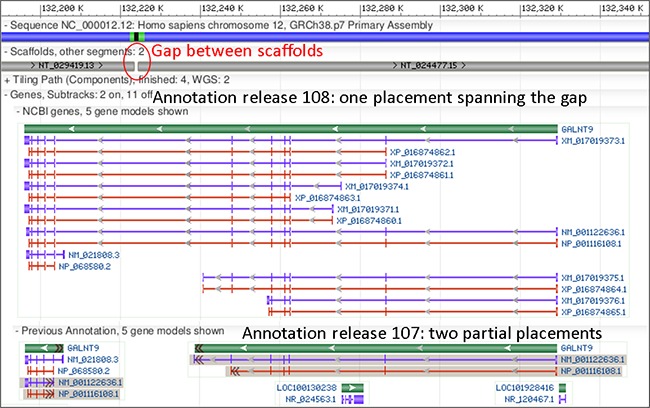
Human genome Annotation Release 108 incorporates new RefSeq sequences, predicts new variants
Friday, June 24, 2016
The Eukaryotic Genome Annotation Pipeline recently produced an updated annotation of the human genome. This new annotation, available in RefSeq, incorporates new "known" RefSeq sequences and predicts new alternative variants. For the first time, this annotation was performed on the assemblies' top-level sequences (chromosomes and unlocalized and unplaced scaffolds). See our previous announcement for details about this change.
Noteworthy improvements in Annotation Release 108 include:
- RNA-Seq datasets used: The Human Protein Atlas (PRJEB4337) and new projects, including the multi-tissue PRJNA280600 and fetal development project PRJNA270632
- 11% increase in coding transcripts for GRCh38.p7 (4,424 known RefSeq and 6,473 model RefSeq)
- 9% increase in non-coding transcripts (1,828 known RefSeq and 2,096 model RefSeq)
Figure 1. Annotation of GALNT9 in Annotation Releases 108 and 107
See Annotation Release 108 in Gene, BLAST, or download it via FTP.
You can find all annotated organisms on the Eukaryotic Genome Annotation Pipeline page.
GenBank release 214.0 is now available via FTP
Thursday, June 23, 2016
GenBank release 214.0 (06/14/2016) has 194,463,572 traditional records containing 213,200,907,819 base pairs of sequence data. In addition, there are 350,278,081 WGS records containing 1,556,175,944,648 base pairs of sequence data, as well as 104,677,061 TSA records containing 94,413,958,919 base pairs of sequence data.
During the 61 days between the close dates for GenBank releases 213.0 and 214.0, the traditional portion of GenBank grew by 1,776,995,772 base pairs and by 724,061 sequence records. During the same period, 313,786 records were updated at an average of 17,013 traditional records added and/or updated per day.
Between releases 213.0 and 214.0, the WGS component of GenBank grew by 103,968,239,699 base pairs and by 11,353,544 sequence records. The TSA component of GenBank grew by 6,602,795,243 base pairs and by 6,529,495 sequence records.
The total number of sequence data files increased by 32 with this release. The divisions are as follows:
- BCT: 11 new files, now a total of 249
- CON: 10 new files, now a total of 344
- ENV: 1 new file, now a total of 92
- INV: 5 new files, now a total of 141
- PAT: 1 new file, now a total of 252
- PLN: 1 new file, now a total of 126
- VRL: 2 new files, now a total of 42
- VRT: 1 new file, now a total of 61
For downloading purposes, please keep in mind that the uncompressed GenBank flat files require approximately 778 GB (sequence files only); the ASN.1 data require approximately 641 GB.
More information about GenBank release 214.0 is available in the release notes.
June 29th webinar: Downloading Exon and Coding Region Sequences for Genes
Monday, June 20, 2016
Next Wednesday, June 29th, NCBI staff will show you how to use the Gene Table report and Graphical Viewer to retrieve exon sequences for genes.
Date and time: Wednesday, June 29, 2016 12:30 PM EDT
Registration: https://attendee.gotowebinar.com/register/859302797289474817
You will also see how to retrieve all exon sequences at once and for multiple genes using the EDirect command line interface to the Entrez search and retrieval system.
After registering, you will receive a confirmation email with information about attending the webinar. After the live presentation, the webinar will be uploaded to the NCBI YouTube channel. Any related materials will be accessible on the Webinars and Courses page; you can also learn about future webinars on this page.
International HapMap Browser to go offline June 16, 2016
Thursday, June 16, 2016
Due to computer security flaws within the HapMap site, it has been decommissioned as of today, June 16, 2016. We regret any inconvenience this sudden removal may cause, but we are acting quickly to ensure security.
The archived HapMap data will continue to be available via FTP. Users can also access the latest data for HapMap samples from the NCBI 1000 Genomes Browser.
See also: NCBI Variation - NCBI retiring HapMap Resource.
NCBI to hold hackathon on NIH campus in August
Tuesday, June 14, 2016
The NCBI and several NIH institutes will host a biomedical data science hackathon at the National Library of Medicine from August 15th to 17th. To apply for this hackathon, complete this form (approximately 10 minutes to complete). Applications are due July 11th, by 4PM ET.
This hackathon will primarily focus on advance bioinformatics analysis of next-generation sequencing data and metadata.
This event is for students, postdocs and investigators or other researchers already engaged in the use of pipelines for genomic analyses from next-generation sequencing data or metadata; it is open to anyone selected for the hackathon who can travel to NIH.
Organization
Participants will be grouped into five to seven teams. These teams will build pipelines and tools to analyze large datasets within a cloud infrastructure.
The potential subjects for this iteration are:
- Calling CNVs
- Parsing large scale bacterial samples
- Statistical analysis of variants mapped to biological networks
- Integration of TCGA and dbGaP metadata
- HL-7 compliance of myfamilyhealthportrait
- Incorporating event detection
- Infectious disease phylogenies
Please see the application for specific team projects.
After a brief organizational session, teams will spend three days analyzing a challenging set of scientific problems related to a group of datasets. Participants will analyze and combine datasets in order to work on these problems.
Datasets
Datasets will come from the public repositories, primarily those housed at the NCBI. During the course, participants will have an opportunity to include other datasets and tools for analysis.
Please note, if you use your own data during the course, we ask that you submit it to a public database within six months of the end of the event.
Products
All pipelines and other scripts, software and programs generated in this course will be added to a public GitHub repository designed for that purpose.
A manuscript outlining the design and usage of the software tools constructed by each team may be submitted to an appropriate journal, such as the F1000Research Hackathons channel.
Application
To apply, complete this form (approximately 10 minutes to complete). Applications are due July 11th, 2016 by 4 pm ET.
Participants will be selected from a pool of applicants based on the experience and motivation they provide on the form. Prior participants and applicants are especially encouraged to reapply.
The first round of accepted applicants will be notified on July 15th by 5 pm ET, and have until July 18th at 9 am to confirm their participation.
If you confirm, please make sure it is highly likely you can attend, as confirming and not attending bars other data scientists from attending this event.
Please include a monitored email address, in case there are follow-up questions.
Notes
Participants will need to bring their own laptop to this program.
A working knowledge of scripting (e.g., Shell, Python) is necessary to be successful in this event. Employment of higher level scripting or programming languages may also be useful.
Applicants must be willing to commit to all three days of the event.
No financial support for travel, lodging or meals is available for this event.
Also note that the course may extend into the evening hours on Monday and/or Tuesday. Please make any necessary arrangements to accommodate this possibility.
Please contact vog.hin@ybsub.neb with any questions.
BLAST+ 2.4.0 now available
Monday, June 13, 2016
Version 2.4.0 of the BLAST+ executables offers improved scoring for selenocysteine residues in the query and database sequences, as well as improved performance for the BLASTP/BLASTX/BLASTN programs.
Download the newest version on the NCBI FTP site. A full list of updates is available in the release notes.
NCBI will transition to HTTPS on September 30, 2016
Friday, June 10, 2016
Starting on September 30th, when you visit NCBI pages, you’ll see a green lock and https:// in the address bar instead of http://. This lets you know that you are really on an NCBI page – that our server identity is confirmed – and that your communication with our server is encrypted and private.
Here’s what to expect if you’re a general user or a scripter:
For general users
You will see the changes mentioned above – https:// and a green lock in the address bar – but you don’t have to update or change anything.
You don’t need to clear your cache or update any links to NCBI pages that you’ve put on your own webpages or shared with people. We will redirect all our pages to https://.
For scripters
To keep calls from failing, use https:, not http:.
Scripts that use HTTP POST to send data will not work once we transition from HTTP to HTTPS on September 30th.
If you'd like to know more about this change to HTTPS, please read The HTTPS-Only Standard from the Federal Chief Information Officers website.
Note: This story originally gave the transition date as September 1, 2016. It has been updated to give the correct date, September 30, 2016. We regret the error.
Tree Viewer 1.9 visualizes medium-large phylogenetic trees
Friday, June 10, 2016
The latest version of Tree Viewer can now visualize medium-large phylogenetic trees up to 15,000 nodes. Tree Viewer 1.9 also includes mini URLs in Link to View, and several other improvements and bug fixes. The Tree Viewer release notes list all updates.
NCBI Tree Viewer is a tool for viewing your own phylogenetic tree data.
June 10th webinar: Finding Systematic Reviews at PubMed Health and PubMed
Tuesday, June 07, 2016
This Friday, NCBI will present a brief instructional webinar that will show you how to find systematic reviews using PubMed and PubMed Health.
Date and time: Friday, June 10, 2016 12:00-12:30 PM EDT
Register here: https://nih.webex.com/nih/k2/j.php?MTID=tfa9ac8a2a377f6cece016f8dd1c9f6f5
After registering, you will receive a confirmation email with information about attending the webinar.
MutaBind: Evaluating the effects of sequence variants and disease mutations on protein-protein interactions
Friday, June 03, 2016
MutaBind is a new computational method and server created through NCBI research efforts that maps mutations on a protein structural complex, calculates changes in binding affinity, identifies deleterious mutations and produces a downloadable mutant structural model.
MutaBind guides you through this process, step by step, starting with selecting a protein complex by inputting PDB code or uploading PDB files. You can also retrieve results with a job ID number, view help documents, and review the MutaBind method and references.
Browse histones, analyze sequences with revamped HistoneDB 2.0
Wednesday, June 01, 2016
Created through research efforts at NCBI, HistoneDB 2.0 is a totally overhauled histone database that can be used to explore the diversity of histone proteins and their sequence variants in many organisms.
This resource was established to better understand how sequence variation may affect functional and structural features of nucleosomes. HistoneDB 2.0 includes distinctive sequence alignments of many histone variants, including H2AZ, macroH2A, subH2B, spermH2B, cenH3, and H3.3, as well as canonical histones.
June 15th webinar: Using NCBI Resources and Variant Interpretation Tools for the Clinical Community
Wednesday, June 01, 2016
In two weeks, NCBI will present a webinar that will show you how to use three clinical variant interpretation tools geared to clinicians through an overview of NCBI variation and medical genetics databases. A demonstration using a clinical case to demonstrate a phenotype-driven whole-genome sequence analysis using tools from Golden Helix, Omicia and SimulConsult will follow the overview.
Date and time: Wednesday, June 15, 2016 1:00 PM EST
Registration URL: https://attendee.gotowebinar.com/register/6974809559951440644
After registering, you will receive a confirmation email with information about attending the webinar. After the live presentation, the webinar will be uploaded to the NCBI YouTube channel. Any related materials will be accessible on the Webinars and Courses page; you can also learn about future webinars on this page.
- Genome Workbench 2.10.7 now available
- July 6th NCBI Minute: Quickly Find Coding Sequences Using ORFfinder
- Human genome Annotation Release 108 incorporates new RefSeq sequences, predicts new variants
- GenBank release 214.0 is now available via FTP
- June 29th webinar: Downloading Exon and Coding Region Sequences for Genes
- International HapMap Browser to go offline June 16, 2016
- NCBI to hold hackathon on NIH campus in August
- BLAST+ 2.4.0 now available
- NCBI will transition to HTTPS on September 30, 2016
- Tree Viewer 1.9 visualizes medium-large phylogenetic trees
- June 10th webinar: Finding Systematic Reviews at PubMed Health and PubMed
- MutaBind: Evaluating the effects of sequence variants and disease mutations on protein-protein interactions
- Browse histones, analyze sequences with revamped HistoneDB 2.0
- June 15th webinar: Using NCBI Resources and Variant Interpretation Tools for the Clinical Community
- NCBI News, June 2016 - NCBI NewsNCBI News, June 2016 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...