NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
HTTPS at NCBI: Guidance for NCBI web API users
Wednesday, July 27, 2016
As originally announced on June 10, NCBI will be moving all web services to the HTTPS protocol on September 30, 2016. Particularly for API users, this move may disrupt any processes that access NCBI APIs using the HTTP protocol. Please see this document on the Develop action page for a complete discussion of this move and what you need to do, along with some new test servers to help you confirm whether your code will function after the change to HTTPS.
dbSNP build 148 for corn, fruit fly, rice and 8 other organisms available
Tuesday, July 26, 2016
dbSNP build 148 is accessible on the web and via FTP. This release includes data for cat, corn, cow, fruit fly, grape, horse, rice, sorghum, tomato, turkey and zebra finch. Build 148 provides over 446 million submitted variants and 225 million reference variants for 14 organisms. To see complete build statistics, visit the SNP summary page.
dbSNP, the NCBI Short Genetic Variations database, catalogs short variations in nucleotide sequences from a wide range of organisms.
August 3rd webinar: NCBI Targeted Loci: RefSeq Ribosomal RNA Sequences for Identification and Phylogenetic Analysis
Thursday, July 21, 2016
On August 3rd, NCBI staff will present a webinar on our targeted loci project. You'll learn about the scope of the project and see practical examples of using these data, BLAST and MOLE-BLAST to identify organisms and explore their diversity in sequences from environmental and organism-associated communities.
Date and time: Wednesday, August 3, 2016 12:00 PM EDT
Registration: https://attendee.gotowebinar.com/register/3690313242319181569
After registering, you will receive a confirmation email with information about attending the webinar. After the live presentation, the webinar will be uploaded to the NCBI YouTube channel. Any related materials will be accessible on the Webinars and Courses page; you can also learn about future webinars on this page.
The targeted loci project is an NCBI-curated set of marker rRNA sequences for prokaryotes (16S, 5S, 23S) and fungi (18S, 28S and ITS). There are now over 27,000 markers representing diverse sets of bacteria, archaea and fungi.
Tree Viewer 1.10 visualizes large phylogenetic trees up to 100,000 nodes
Tuesday, July 19, 2016
Tree Viewer version 1.10 has several improvements, updates and bug fixes, including the ability to visualize large phylogenetic trees up to 100,000 nodes. In addition, Tree Viewer 1.10 has added functionality for sorting, as well as improved API and zoom functions. The Tree Viewer release notes list all updates.
NCBI Tree Viewer is a tool for viewing your own phylogenetic tree data.
NCBI Insights blog post: NCBI is Phasing out Sequence GIs - Here's What You Need to Know
Friday, July 15, 2016
The latest blog post on NCBI Insights answers two questions you may have regarding the switch to accession.version:
- What pieces of your code will break in September?
- Are GI numbers gone for good?
NCBI Insights is the official NCBI blog, where we share science feature stories, quick tips and what’s new at NCBI.
July 27th NCBI Minute: Important Changes to NCBI Web Protocols
Wednesday, July 13, 2016
In two weeks, we’ll discuss NCBI’s upcoming switch to the secure HTTPS protocol. Through this NCBI Minute, you’ll learn how this change will affect your access to NCBI pages and services and what you should do to have a smooth transition.
Date and time: Wednesday, July 27, 2016 12:00 PM EDT
Registration URL: https://attendee.gotowebinar.com/register/2000297899730334722
After registering, you will receive a confirmation email with information about attending the webinar. After the live presentation, the webinar will be uploaded to the NCBI YouTube channel. Any related materials will be accessible on the Webinars and Courses page; you can also learn about future webinars on this page.
Sequence Viewer 3.15 is now available
Wednesday, July 13, 2016
Sequence Viewer 3.15 brings several new features, improvements and bug fixes to the graphical viewer, including improved PDF graphics for markers, an extended embedding API and improved tooltips. For a full list of changes, see the release notes.
Sequence Viewer is a graphical view of sequences and color-coded annotations on regions of sequences stored in the Nucleotide and Protein databases.
July 20th NCBI Minute: Important Changes Coming to Sequence Databases
Tuesday, July 12, 2016
In next Wednesday's NCBI Minute, NCBI staff will describe upcoming changes to sequence records. Starting this September, GI identifiers will no longer appear on GenBank and FASTA record views; we will discuss the details and consequences of this change, along with the future of existing GI identifiers.
Date and time: Wednesday, July 20, 2016 12:00 PM EDT
Registration URL: https://attendee.gotowebinar.com/register/6741098469079346177
After registering, you will receive a confirmation email with information about attending the webinar. After the live presentation, the webinar will be uploaded to the NCBI YouTube channel. Any related materials will be accessible on the Webinars and Courses page; you can also learn about future webinars on this page.
Conserved Domain Database (CDD) version 3.15 now available online and via FTP
Tuesday, July 12, 2016
Version 3.15 of the Conserved Domain Database contains 290 new or updated NCBI-curated domains (52,411 total), including models specifically built to annotate structural motifs (accession prefix "sd"), and now mirrors Pfam version 28.
Updates include:
- Fine-grained classification of the beta lactamase-like metallohydrolases
- Conserved domain hits in CD Search are ranked by E-value, without giving preference to NCBI-curated models.
You can access CDD at the Conserved Domains homepage and find updated content on the CDD FTP site.
RefSeq release 77 is now available
Thursday, July 07, 2016
RefSeq release 77 is accessible online, via FTP and through NCBI’S programming utilities. This full release incorporates genomic, transcript and protein data available as of June 29, 2016 and includes:
- 100,678,438 records,
- 65,964,245 proteins,
- 15,563,994 RNAs,
- and sequences from 60,892 organisms.
The release is provided in several directories as a complete dataset and also as divided by logical groupings.
Additional information about release 77 can be found in the release notes. You can also directly receive statistics and a summary of announcements for each RefSeq release by subscribing to the refseq-announce listserv.
Mouse and zebrafish genome annotations updated
Wednesday, July 06, 2016
The mouse (GRCm38.p4) and zebrafish (GRCz10) genomes were recently re-annotated by the Eukaryotic Genome Annotation Pipeline. For both, the annotation was performed on the RefSeq assemblies' top-level sequences (chromosomes and unlocalized and unplaced scaffolds). See our previous announcement for details about this change.
Mouse (mus musculus)
The new annotation includes 2,378 "known" RefSeq transcripts that are new or were modified since the previous release. The proportion of protein-coding genes represented by at least one "known" RefSeq is now 91%.
See Mus musculus Annotation Release 106 in Gene, BLAST, or via download.
Zebrafish (Danio rerio)
The new annotation includes 2,364 annotated "known" RefSeq transcripts that are new or were modified since the previous release. The proportion of protein-coding genes represented by at least one "known" RefSeq is now 55%.
See Danio rerio Annotation Release 105 in Gene, BLAST, or via download.
Note: Genes spanning adjacent scaffolds may now be represented as a single feature. See, for example, mouse genes Dock2, Rims1, or Immp2l and zebrafish nduaf11.
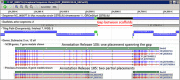
Figure
Figure 1. Annotation of mouse Dock2 in Annotation Release 106 and 105
You can find all annotated organisms on the Eukaryotic Genome Annotation Pipeline page.
- HTTPS at NCBI: Guidance for NCBI web API users
- dbSNP build 148 for corn, fruit fly, rice and 8 other organisms available
- August 3rd webinar: NCBI Targeted Loci: RefSeq Ribosomal RNA Sequences for Identification and Phylogenetic Analysis
- Tree Viewer 1.10 visualizes large phylogenetic trees up to 100,000 nodes
- NCBI Insights blog post: NCBI is Phasing out Sequence GIs - Here's What You Need to Know
- July 27th NCBI Minute: Important Changes to NCBI Web Protocols
- Sequence Viewer 3.15 is now available
- July 20th NCBI Minute: Important Changes Coming to Sequence Databases
- Conserved Domain Database (CDD) version 3.15 now available online and via FTP
- RefSeq release 77 is now available
- Mouse and zebrafish genome annotations updated
- NCBI News, July 2016 - NCBI NewsNCBI News, July 2016 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...
