NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Sequence Viewer 3.20 is now available
Thursday, March 30, 2017
Sequence Viewer 3.20 has several new features, improvements and bug fixes, including discrete color maps for graph tracks, improved performance in initialization and loading tracks, improved display of overlapping variation features and the addition of a status bar. For a full list of changes, see the Sequence Viewer release notes.
Sequence Viewer is a graphical view of sequences and color-coded annotations on regions of sequences stored in the Nucleotide and Protein databases.
Conserved Domain Database (CDD) version 3.16 now available online and via FTP
Thursday, March 30, 2017
Version 3.16 of the Conserved Domain Database contains 1,659 new or updated NCBI-curated domains (56,066 total), including models specifically built to annotate structural motifs (accession prefix "sd"), and now mirrors Pfam version 30.
Updates include:
- Fine-grained classification of the 7-membrane GPCR transmembrane subunits.
- Database size parameters for CD-Search have been adjusted, resulting in slightly higher E-values.
- Fewer models are now assigned a multi-domain-model status, affecting the domain annotation of a large number of proteins.
You can access CDD at the Conserved Domains homepage and find updated content on the CDD FTP site.
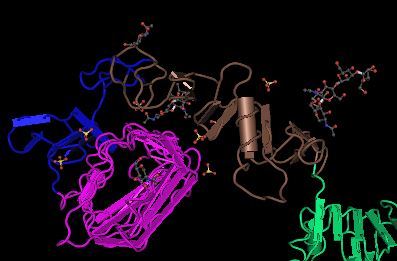
Type 1 Insulin-like Growth Factor Receptor (1IGR), colored by domain.
NCBI to assist with BioFrontiers Hackathon in May
Monday, March 27, 2017
From May 22nd to 24th, NCBI will be assisting with the BioFrontiers Hackathon in Boulder, Colorado. Please see the BioFrontiers Hackathon website for more information, including what to expect, who should apply, and the application itself. Applications are due by April 7, 2017.
NCBI will attend the AACR Annual Meeting 2017
Tuesday, March 21, 2017
From April 2-5, 2017, NCBI will attend the American Association for Cancer Research (AACR) Annual Meeting in Washington, DC. Join us at Exhibit Booth #3230 at the following times:
- Sunday, April 2, 1 – 5pm
- Monday, April 3, 9am – 5pm
- Tuesday, April 4, 9am – 5pm
- Wednesday, April 5, 9am – 12 noon
At the booth, you'll be able to have your questions answered and see demos of NCBI resources pertaining to medical genetics, sequences and their variations, and biomedical literature.
Genome Workbench 2.11.10 now available
Monday, March 20, 2017
The latest version of Genome Workbench includes a number of new features, fixes and improvements like a critical improvement in HTTPS protocol communication with NCBI and a new coloring scheme in Multiple Alignment View.
For a full list of changes, please see the Genome Workbench release notes.
Tree Viewer version 1.13 implements new search in tree features
Monday, March 20, 2017
Tree Viewer version 1.13 has several improvements, updates and bug fixes, including search in trees, improved automatic subtree collapse, and more. The Tree Viewer release notes list all updates.
NCBI Tree Viewer is a tool for viewing your own phylogenetic tree data.
Complete RefSeq genome annotation results represented in UCSC genome browser
Friday, March 17, 2017
We are very pleased to announce the availability of the complete RefSeq human genome annotation product for the GRCh38 assembly in the University of California, Santa Cruz (UCSC) Genome browser. NCBI and UCSC staff have worked closely to define an improved data exchange process and NCBI is now providing RefSeq genome annotation and alignment data in order to have a more complete reflection of the RefSeq product in the UCSC genome browser. This resolves issues of incomplete data and conflicting placement details between UCSC displays and NCBI displays.
This initial release is for the human reference genome (GRCh38) and does not include NCBI RefSeq annotation for GRCh38 patches added since the initial GRCh38 release. We anticipate working with UCSC to expand on the number of organisms in the future.
NCBI-provided RefSeq data is included in the "NCBI RefSeq" composite track. For the following tracks, the alignments and coordinates are provided by RefSeq:
- RefSeq All – curated and predicted transcript annotations
- RefSeq Curated – curated annotations (transcripts with NM_ and NR_ accessions)
- RefSeq Predicted – predicted annotations (transcripts with XM_ and XR_ accessions)
- RefSeq Other –annotations not included in RefSeq All such as pseudogenes or other loci
- RefSeq Alignments – alignments of transcripts to the genome provided by RefSeq
By default, only the "RefSeq Curated" subtrack is activated within the "NCBI RefSeq" track, but you may wish to activate the other subtracks to view the complete dataset.
March 29th NCBI Minute: How to Submit Your 16S rRNA Data to NCBI
Friday, March 17, 2017
In two weeks, NCBI staff will guide you through the submission of prokaryotic 16S rRNA sequences to GenBank using one of the new Submission Wizards.
Date and time: Wednesday, March 29, 2017 12:00 PM - 12:30 PM EDT
After registering, you will receive a confirmation email with information about attending the webinar. After the live presentation, the webinar will be uploaded to the NCBI YouTube channel. Any related materials will be accessible on the Webinars and Courses page; you can also learn about future webinars on this page.
NCBI will attend the 2017 Annual Clinical Genetics Meeting
Wednesday, March 15, 2017
Join NCBI staff at the 2017 Annual Clinical Genetics Meeting (ACMG) in Phoenix, AZ on March 21-25, 2017. At Exhibit Booth #531, you’ll be able to get navigation tips, hands-on help, and handout materials.
In addition, Adriana Malheiro will present NCBI's suite of human genome resources that support the Precision Medicine Initiative in a Platform Presentation titled "In the Clinic with Medical Genetics Summaries (MGS)".
Finally, Melissa Landrum and Adriana Malheiro will present posters titled "ClinVar: For medical practitioners and researchers alike" and "MedGen: Harmonizing phenotypic information into an online, computer-readable resource of medical genetics", respectively.
Please see the NCBI Conferences & Presentations page, as well as the official NCBI and NCBI Clinical Twitter accounts once ACMG starts for further information about presentations and posters, as well as times and locations.
RefSeq release 81 now public
Tuesday, March 14, 2017
RefSeq release 81 is now accessible online, via FTP and through NCBI's programming utilities. This full release incorporates genomic, transcript, and protein data available as of March 6, 2017 and contains 121,954,847 records, including 81,027,309 proteins, 18,381,587 RNAs, and sequences from 68,165 organisms. The release is provided in several directories as a complete dataset and also as divided by logical groupings.
GI sequence identifiers removed from flatfile and FASTA formats
Please refer to these NCBI News announcements for more details:
Comprehensive reannotation of prokaryotic genomes
The first phase of the comprehensive reannotation of prokaryotic genomes has been completed, covering Escherichia, Shigella, Salmonella, Klebsiella, and Listeria.
Reannotation is expected to be completed before the May 2017 RefSeq release.
Information on the improvements to the Prokaryotic Genome Annotation Pipeline 4.1 can be found here.
FTP files for RefSeq prokaryote genomes on the genomes FTP site will be refreshed upon completion of the reannotation project.
GenBank release 218.0 is now available
Tuesday, March 14, 2017
GenBank release 218.0 (2/13/2017) has 199,341,377 traditional records containing 228,719,437,638 base pairs of sequence data. In addition, there are 409,490,397 WGS records containing 1,892,966,308,635 base pairs of sequence data, 151,431,485 TSA records containing 133,517,212,104 base pairs of sequence data, as well as 1,438,349 TLS records containing 636,923,295 base pairs of sequence data.
During the 60 days between the close dates for GenBank releases 217.0 and 218.0, the traditional portion of GenBank grew by 3,746,377,205 base pairs and by 775,902 sequence records. During the same period, 68,617 records were updated at an average of 14,075 traditional records added and/or updated per day.
Between releases 217.0 and 218.0, the WGS component of GenBank grew by 75,776,742,790 base pairs and by 14,189,221 sequence records. The TSA component of GenBank grew by 8,188,387,596 base pairs and by 9,337,148 sequence records. The TLS component of GenBank grew by 52,225,376 base pairs and by 169,659 sequence records.
The total number of sequence data files increased by 39 with this release. The divisions are as follows:
- BCT: 26 new files, now a total of 330
- CON: 1 less file, now a total of 356
- ENV: 1 new file, now a total of 95
- INV: 1 new file, now a total of 152
- PAT: 3 new files, now a total of 283
- PLN: 6 new files, now a total of 143
- VRL: 2 new files, now a total of 47
- VRT: 1 new file, now a total of 64
For downloading purposes, please keep in mind that the uncompressed GenBank Release 218.0 flatfiles require approximately 818 GB (sequence files only); the ASN.1 data require approximately 677 GB.
More information about GenBank release 218.0 is available in the release notes, as well as in the README files in the genbank (ftp.ncbi.nih.gov) and ASN.1 (ncbi-asn1) directories.
Seven new annotations added to RefSeq
Thursday, March 09, 2017
In the past month, the NCBI Eukaryotic Genome Annotation Pipeline has released new annotations in RefSeq for the following organisms:
- Asparagus officinalis (garden asparagus)
- Microcebus murinus (gray mouse lemur)
- Aegilops tauschii (a monocot)
- Cajanus cajan (pigeon pea)
- Castor canadensis (American beaver)
- Ananas comosus (pineapple)
- Paralichthys olivaceus (Japanese flounder)
See more details on the Eukaryotic RefSeq Genome Annotation Status page.
Multiple Sequence Alignment Viewer 1.4 is now available
Thursday, March 09, 2017
The new version of the Multiple Sequence Alignment Viewer (MSA Viewer) has implemented several bug fixes affecting several features, including zoom on alignments and text import. A full list of bug fixes is available in the MSA Viewer release notes.
Magic-BLAST 1.2.0 now available
Monday, March 06, 2017
The newest version of Magic-BLAST handles multiple SRA accessions, offers improved splice site detection and multi-threading performance, and fixes issues with macOS installation. For more information, see the release notes. The new executables are available on the NCBI FTP site.
Magic-BLAST is a tool for mapping large next-generation RNA or DNA sequencing runs against a whole genome or transcriptome. Read more here.
Expression section and bulk datasets added to NCBI Gene
Monday, March 06, 2017
The Gene resource has a new feature that reports normalized RNA expression levels computed from RNA-Seq data for human, mouse, and rat genes. An expression chart is available on the Gene full report pages, with an additional table view and download option on the new expression report page available through the “See details” link or format menu.
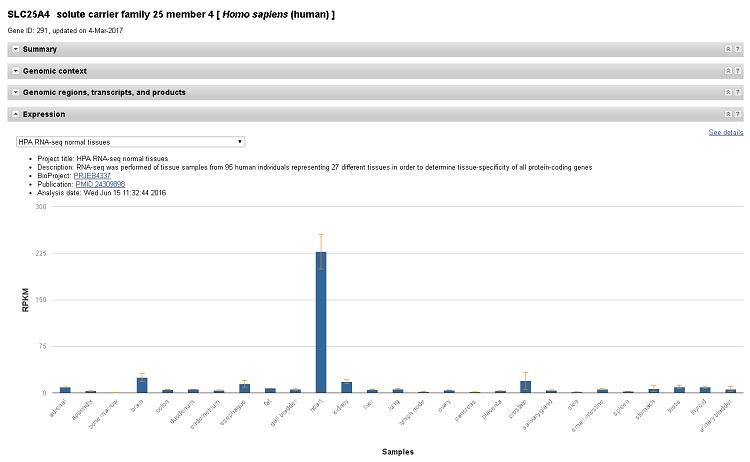
Figure 1. The new Expression section on Gene pages.
Expression data can provide key insights into where and when a gene may be functioning, for example by exposing the correlation between expression of human SLC25A4 and its established role in heart function.
Bulk datasets will also be available on the Gene FTP site. The RNA-Seq expression coverage graphs for each sample used to compute expression levels are available in the embedded graphical viewer and Genome Data Viewer under the expression category. We welcome questions about this new dataset at vog.hin.mln.ibcn@ofni or through the "Contact Help Desk" link available on the Gene full report page.
- Sequence Viewer 3.20 is now available
- Conserved Domain Database (CDD) version 3.16 now available online and via FTP
- NCBI to assist with BioFrontiers Hackathon in May
- NCBI will attend the AACR Annual Meeting 2017
- Genome Workbench 2.11.10 now available
- Tree Viewer version 1.13 implements new search in tree features
- Complete RefSeq genome annotation results represented in UCSC genome browser
- March 29th NCBI Minute: How to Submit Your 16S rRNA Data to NCBI
- NCBI will attend the 2017 Annual Clinical Genetics Meeting
- RefSeq release 81 now public
- GenBank release 218.0 is now available
- Seven new annotations added to RefSeq
- Multiple Sequence Alignment Viewer 1.4 is now available
- Magic-BLAST 1.2.0 now available
- Expression section and bulk datasets added to NCBI Gene
- NCBI News, March 2017 - NCBI NewsNCBI News, March 2017 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...