NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Phase One Rollout of the New Genome Site
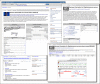
A completely redesigned Genome site, www.ncbi.nlm.nih.gov/genome, is now available. Major improvements include a more natural organization at the level of the organism for prokaryotic, eukaryotic, and viral genomes. Reports include information about the availability of nuclear or prokaryotic primary genomes as well as organelles and plasmids. The new Genome resource provides a summary view of the data from all genome-scale projects including genome maps, assemblies, annotation, and transcriptomes. Genome collects data from primary data resources and provides links to more detailed information. While not new with this release, it is worth noting that the Genome interface has been upgraded to the new NCBI standard with the new search bar, Limits and Advanced Search pages, and NCBI footer. Moreover, search results and record views in Genome are discovery-oriented and feature the Discovery Column with analysis tools and easy access to related data. Figure 1 shows sample pages from the Genome resource and highlights these new features. An information page accompanying the release provides additional details and help with transitioning to the new service.
The new Genome site is much easier to navigate and provides rapid access to all genome data for a particular organism. The new site will continue to improve as additional displays and features are added in phases. A feature article in the next NCBI News will provide more detailed coverage of Genome with examples illustrating the power of the new system.
Note: Changes affecting genome identifiers
Because of the reorganization to a natural classification system, older genome identifiers are no longer valid. Typically these genome identifiers were not exposed in the previous system and were used mainly for programmatic access. To aid in the transition to the new system, a file that maps previous Genome identifiers to the identifier for the genome sequence is available on the FTP site. The sequence identifiers can be used to retrieve the genome sequence from the Nucleotide database.
New BLAST videos on NCBI's YouTube channel
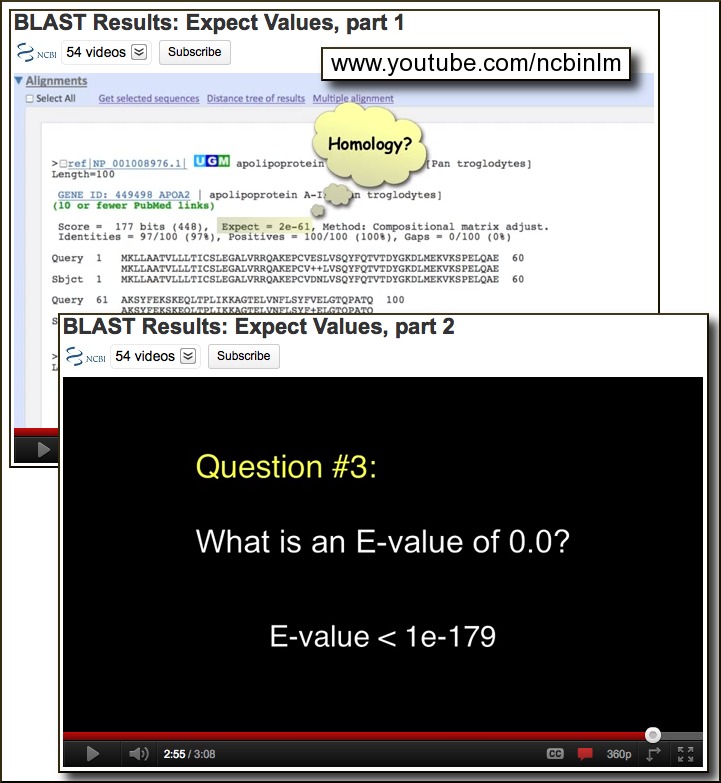
Two new instructional videos about BLAST statistics are on the NCBI YouTube channel: An explanation of BLAST E-values (pt.1), and Answers to a few E-value FAQs (pt.2). These BLAST videos should help in interpreting BLAST output and designing more effective BLAST search strategies. The BLAST videos join a growing collection of 54 videos on the NCBI YouTube channel.
Entrez Utility Changes: New EFetch Version and New alternative ESummary XML
An updated EFetch Entrez Utility (version 2.0) is now in production. EFetch retrieves records from the NCBI databases by unique identifier. Additions include support for the BioSample , BioProjects, and SRA databases as well as defined default values for retrieval mode (retmode) and retrieval type (rettype). Updated retmode and rettype values are given in the table in the Entrez Programming Utilities Help Manual.
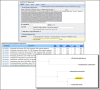
An alternative XML record is now available from the ESummary Entrez Utility. The content in the new record is unique to each Entrez database and has additional content not available in the traditional ESummary record. The new XML can be requested by including &version=2.0 in the ESummary URL. The traditional ESummary record will continue to be supported and will be returned without the version parameter. The traditional and version 2.0 ESummary for NM_000240 (gi=33469954) from the nuccore (nucleotide) database are shown below. The release notes provide details on changes. The Entrez Programming Utilities Help Manual has complete information on EFetch, ESummary, and the other EUtility programs.

Highlight Features Link Now on Sequence Records
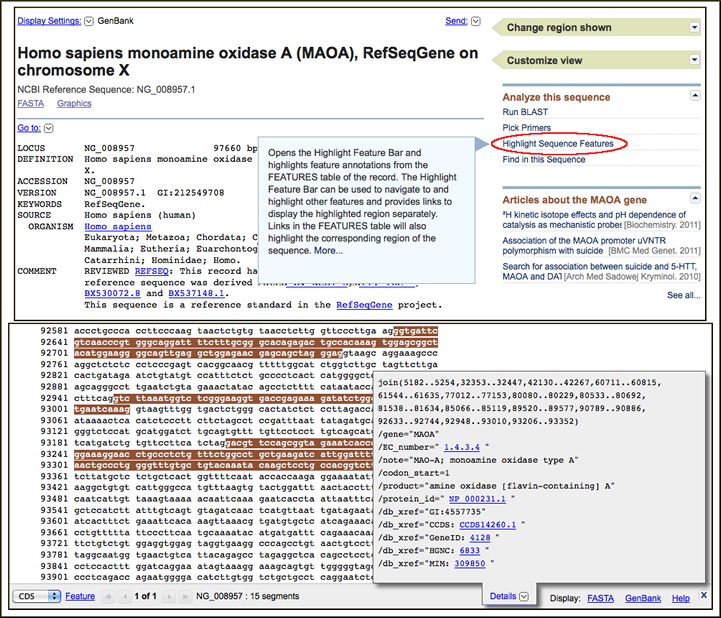
A Highlight Features link now appears in the Analyze this Sequence section of protein and nucleotide sequence records displayed at the NCBI site. This link activates the new Feature Highlight function described in the August 2011 NCBI News. Clicking the link opens the Feature Highlight Bar and highlights the first coding sequence (CDS) feature or the first linked feature if no CDS feature is present.
As pointed out in the original NCBI News article, the Highlight Features function is helpful in visualizing the extent and location of such important features as genes, coding regions, exons, and mRNAs in nucleotide sequences and conserved domains, modification sites, and interaction sites in protein sequences. This function joins the links to BLAST, Primer-BLAST, and Conserved Domain searches as well as the Find-in-Sequence pattern finder as on-the-fly analysis capabilities in the NCBI sequence databases. The image below shows the link and the activated CDS highlight on the ReSeqGene record for the human monoamine oxidase gene (NG_008957).
New BLAST 16S Prokaryotic Ribosomal RNA Database
A prokaryotic 16S ribosomal RNA database is now available through the database pull-down list on the main nucleotide BLAST service. The 16S database contains both bacterial and archaeal sequences from two RefSeq Targeted Loci projects (BioProjects PRJNA33175 and PRJNA33317). These data represent near full-length 16S ribosomal RNA sequences from more than 250 archaeal and 7200 bacterial strains. The 16S BLAST database is useful for identifying or establishing the taxonomic affinities of unknown bacterial 16S sequences such as those from environmental or organismal samples or metagenomes. Figure 2 shows how the database can be used to partially classify a 16S sequence (JF340503) obtained from a concrete sewer biofilm (PubMed: 21981064, PopSet: 330372088). The top panel of the fgure shows the basic nucleotide BLAST form with the 16S database selected. The center panel shows the BLAST results (RID: CUR81JZY012). The results indicate that the query sequence has the closest affinity to the acetobacteriaceae, particularly Acidocella facilis. The BLAST Distance Tree, also shown in the figure provides a useful way to see the results of the analysis at a glance.
The pre-formatted16S microbial database is also available in the BLAST db FTP directory as the file 16SMicrobial.tar.gz.
New Phenotype-Genotype Integrator (PheGenI)
The Phenotype-Genotype Integrator (PheGenI) is a new service that integrates genome-wide association study (GWAS) catalog data from NHGRI with molecular and literature databases at the NCBI. PheGenI takes chromosome location, gene, SNP, or phenotype as input and provides annotated tables of SNPs, genes, association results, and gene expression data. A new tutorial video on YouTube demonstrates how to use PheGenI.
Eukaryotic Genome Builds and Updates
Twelve new genome assemblies with annotations have recently been released at the NCBI. Nine of the new builds are genomes that make their first appearance (build 1.1) at NCBI. Highlights include the first genome for a sponge (Amphimedon queenslandica), the first for a reptile – the green anole (Anolis carlonensis), the first for a perciform fish – the Nile tilapia (Oreochromis niloticus), and two new rodent genomes – the guinea pig (Cavia porcellus) and the Chinese hamster CHO-K1 cell line (Cricetulus griseus). In addition updated annotations for six more genomes are also available including human build 37.3 described in the next section. A complete list of new builds and updates is given below. The NCBI BioProject, Genome, Gene, Nucleotide, Protein, BLAST and Map Viewer services provide access to these data. The assemblies and annotations may also be downloaded from the genomes area of the FTP site.
First NCBI Builds (build 1.1)
Sponge (Amphimedon queenslandica) [BioProject, Map Viewer]
Buff-tailed bumblebee (Bombus terrestris) [BioProject, Map Viewer]
Nile tilapia (Oreochromis niloticus) [BioProject, Map Viewer]
Domestic turkey (Meleagris gallopavo) [BioProject, Map Viewer]
Green Anole (Anolis carolinensis) [BioProject, Map Viewer]
Guinea pig (Cavia porcellus) [BioProject, Map Viewer]
African savannah elephant (Loxodonta africana) [BioProject, Map Viewer]
White-faced gibbon (Nomascus leucogenys) [BioProject, Map Viewer]
Chinese hamster (CHO-K1 cell line) (Cricetulus griseus) [BioProject, Map Viewer]
New Builds
Honeybee (Apis mellifera), build 5.1 [BioProject, Map Viewer]
Pea aphid (Acyrthosiphon pisum), build 2.1 [BioProject, Map Viewer]
Zebrafish (Danio rerio), build 5.1 [BioProject, Map Viewer]
Chimpanzee (Pan troglodytes), build 3.1 [BioProject, Map Viewer]
Updated Annotations
Fruit fly (Drosophila melanogaster), build 9.4 [BioProject, Map Viewer]
Horse (Equus caballus), EquCab2.0 [BioProject, Map Viewer]
Dog (Canis lupus familiaris), build 2.2 [BioProject, Map Viewer]
Duck-billed platypus (Ornithorhynchus anatinus), build 1.2 [BioProject, Map Viewer]
Thale cress (Arabidopsis thaliana), TAIR 10 [BioProject, Map Viewer]
Human (Homo sapiens), build 37.3 [BioProject, Map Viewer]
Human Genome Update
The NCBI human genome annotation has been updated to version 37.3 and is now available in the Map Viewer, the Entrez system and human genome BLAST. The build statistics have more information on the contents of the release. The update includes the Genome Reference Consortium sequence patches from patch 5. The patches are currently available as separate sequences from the chromosome assemblies. Patches that correct problems in the current assembly (fix patches) will be incorporated in the next complete genome assembly (build 38).
Microbial Genomes Update
Fifty-eight finished microbial (archaeal and bacterial) genomes were released during September and October 2011. The original sequence data files submitted to the International Sequence Database Collaboration (INSDC) are available in the Bacteria directory in the genomes area of the GenBank FTP site. RefSeq provisional versions were released for a selected set of 32 of the complete INSDC microbial genomes during the same period. These are available from the /genomes/Bacteria directory on the FTP site.
In addition, data from 425 microbial whole genome-shotgun (WGS) sequencing projects were added to the INSDC during this period. The original submitted files are available in the Bacteria_DRAFT directory in the GenBank genomes area. RefSeq provisional versions of 89 WGS microbial projects were released in the /genomes/Bacteria_DRAFT area of the FTP site.
All GenBank and RefSeq microbial genomes are incorporated in the NCBI integrated Entrez search and retrieval system and the BLAST sequence similarity search service.
GenBank News
GenBank release 186 is available through Entrez, BLAST and from the GenBank FTP area. The current release incorporates data available as of Oct 13, 2011 and, with the whole-genome shotgun portion, contains 350,733,781,429 bases from 212,788,863 sequence records. Release notes describe the current state of data and upcoming changes.
RefSeq News
RefSeq Release 50 is available through Entrez, BLAST, and from the RefSeq FTP area. The current release includes 18.8 million Reference Sequence records from 16,392 different species or strains. The RefSeq release notes provide more detailed information.
Conserved Domain Database Update
Version 3.01 of the Conserved Domain Database is now available. The new release contains 298 new or updated NCBI-curated domain models. More detailed statistics are available from the CDD News page. CDD matrices and other information can be downloaded from the FTP site. CDD data are incorporated in the Entrez and BLAST search services at the NCBI Website.
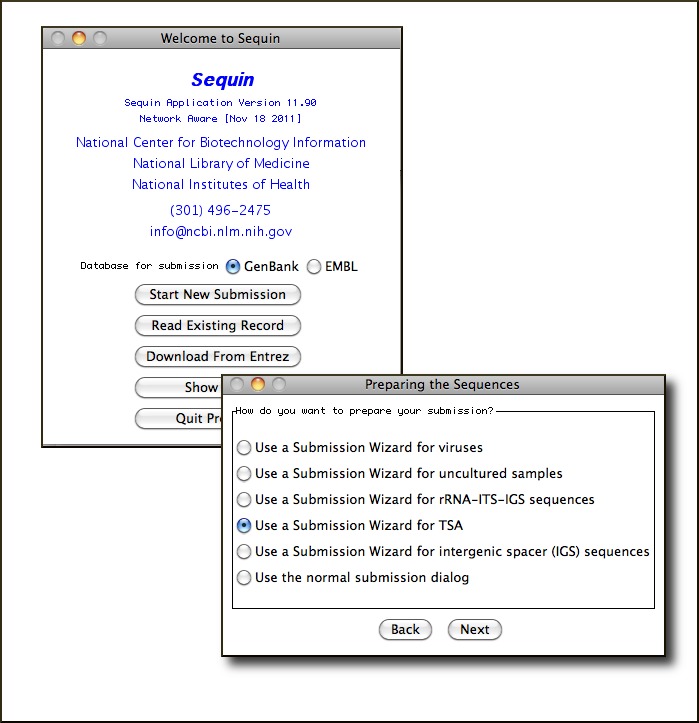
Sequin Now with Transcriptome Shotgun Assembly and Internal Transcribed Spacer Sequence Submission Wizards
A new version (11.90) of Sequin, the NCBI's standalone submission preparation software, is now available for download. Packages are available for Linux, Unix, Windows, and Mac OSX systems. Improvements include new Submissions Wizards for Transcriptome Shotgun Assemblies (TSA) and ribosomal RNA intergenic spacer sequences (ITS); a Sequencing Method Page for information about the sequencing technology and assembly methods; a Sequence Deletion Tool for removing sequences from the submission; and updated feature and qualifier wizards complying with the latest INSDC Feature Documentation. The Sequin page has more information, a Quick guide, FAQs, and extensive help documentation on using Sequin to prepare submissions.
NCBI C++ Toolkit Major Release
NCBI C++ Toolkit v7.0.0 is now available from the FTP site. The release notes describe the highlights and contents of this release. The Toolkit contains C++ language sources of NCBI software that can be used to build standalone BLAST, Sequin, Cn3D, and other NCBI tools and utilities. The NCBI C++ Toolkit Book has in-depth information on working with the toolkit and provides access to source browsers and other useful resources.
Announce Lists and RSS Feeds
Seventeen topic-specific mailing lists are available that provide email announcements about changes and updates to NCBI resources including dbGaP, BLAST, GenBank, and Sequin. The various lists are described on the Announcement List summary page. Subscribe to the NCBI Announce list to receive updates on the NCBI News.
Twenty-one RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce.
NCBI’s Facebook page and Twitter feed also provide updates on NCBI resources.
Send comments and questions about NCBI resources to info@ncbi.nlm.nih.gov, or call 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
- Phase One Rollout of the New Genome Site
- New BLAST videos on NCBI's YouTube channel
- Entrez Utility Changes: New EFetch Version and New alternative ESummary XML
- Highlight Features Link Now on Sequence Records
- New BLAST 16S Prokaryotic Ribosomal RNA Database
- New Phenotype-Genotype Integrator (PheGenI)
- Eukaryotic Genome Builds and Updates
- Human Genome Update
- Microbial Genomes Update
- GenBank News
- RefSeq News
- Conserved Domain Database Update
- Sequin Now with Transcriptome Shotgun Assembly and Internal Transcribed Spacer Sequence Submission Wizards
- NCBI C++ Toolkit Major Release
- Announce Lists and RSS Feeds
- NCBI News, November 2011 - NCBI NewsNCBI News, November 2011 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...