NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Featured Resource: BLAST 2 Sequences Is Now Part of the Main BLAST Web Service
The former BLAST utility program Blast 2 Sequences is now available on all main BLAST Web forms. This program eliminates the need for the separate service formerly linked as a specialized BLAST page. More importantly, the service now provides the full functionality of the Web BLAST engine, the BLAST formatter, and the ability compare more than one sequence in a single search.
Accessing and Using the Service
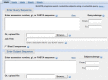
A Blast 2 Sequences checkbox is now available on all BLAST forms linked to the Basic BLAST section of the BLAST homepage (blast.ncbi.nlm.nih.gov). The old Blast 2 Sequences link on the BLAST Homepage will link directly to the Basic Nucleotide form already set up for Blast 2 Sequences. On the BLAST submission page, checking the Blast 2 Sequences checkbox changes the form so that the database selection portion is eliminated, and a new text area appears for entering target sequences (Figure 1). The newly added program tabs allow rapid selection of a different program if desired (blastp, blastn, blastx, tblastn, tblastx). Sequences in FASTA format or database accession numbers may be entered in either of the two text areas. Sets of sequences may also be uploaded to the service from files on disk. The ability to enter multiple sequences makes it possible to search a small custom database on the web or to perform an all-against-all comparison of a small number of sequences.
Output Format
The Blast 2 Sequences output provides a completely redesigned dot plot of the alignment that features a higher resolution display than the old Blast 2 Sequences service. The dot-plot may be collapsed if desired. The dot-plot is useful in many cases for identifying repeated domains in proteins or insertions, inversions, and translocations in nucleotide sequences. Figure 2 shows the dot-plot of the alignment of two bacterial genomes with large-scale relative rearrangements. The rearrangements are easily visible in the plot.
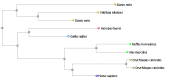
All formatting options available for the main Web BLAST service are also available for Blast 2 Sequences. These options include alternative alignment views such as pairwise, query anchored with mismatch and identity highlighting, and downloadable structured formats such as ASN.1, XML, and hit table. The ability to display results as a distance tree using the BLAST Treeview link is particularly useful when the Blast 2 Sequences option is used with a small custom database as shown in Figure 3 for selected vertebrate aromatic amino acid hydroxylases.
Summary
The Blast 2 Sequences utility is now fully integrated into the main NCBI BLAST service, providing the full power of the Web BLAST service along with flexible formatting options for comparing two sequences. This feature also extends the capability of Blast 2 Sequences to make it a “Blast several Sequences” service. These expanded options and abilities make Blast 2 Sequences a new, powerful, and flexible sequence analysis tool.
New Databases and Tools
PMID : PMCID Converter
A converter is available to translate ID numbers for articles found in both PubMed and PubMed Central. The converter will convert IDs from PubMed to PMC and vice versa. It can be found at: http://www.ncbi.nlm.nih.gov/sites/pmctopmid
Bookshelf
The Bookshelf has added two new books entitled: The Epilepsies: Seizures, Syndromes, and Management and Essentials of Glycobiology. Books can be found at: www.ncbi.nlm.nih.gov/sites/entrez?db=Books.
Genome Resource Guide
An Aphid Genome Resource page is available at: www.ncbi.nlm.nih.gov/projects/genome/guide/aphid/ . This page provides a gateway to aphid resources both within NCBI and the outside scientific community.
Microbial Genomes
Fifteen finished microbial genomes were released from October 10-November 17. The original sequence data files submitted to GenBank/EMBL/DDBJ can be found at: ftp.ncbi.nih.gov/genbank/genomes/Bacteria/. The RefSeq provisional versions of these genomes are available via FTP at: ftp.ncbi.nih.gov/genomes/Bacteria/.
GenBank News
GenBank release 168.0 is available via web and FTP that includes information as of October 27, 2008. The database increased by 19.84 Gigabases since the last release, 167.0. This is a milestone for the largest growth between single releases.
Updates and Enhancements
RefSeq
RefSeq Release 32 is available via web and FTP. This full release incorporates genomic, transcript, and protein data available as of November 7, 2008 and includes 9,145,702 records from 5,513 different organisms. The RefSeq website is: www.ncbi.nlm.nih.gov/RefSeq/.
RefSeq records for microbial projects with the prefix ‘NZ’ will now include two style formats where there may be 2 or 4 alphabetic characters following the underscore. These new accession formats have been designed to support duplicating WGS microbial scaffolds and complete genomic models when a project is a mixture of contigs and scaffolds.
Genome Assembly
Genome annotation for Anopheles gambiae build AgamP3.3 and Caenorhabitis elegans build WS190 were released in October. For more annotation updates and links to Genome Resource pages see: www.ncbi.nlm.nih.gov/Genomes/.
Announce Lists and RSS Feeds
Fifteen topic-specific mailing lists are described on the Announcement List summary page. Announce lists provide email announcements about changes and updates to NCBI resources. www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html
Seven RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce. Please see: www.ncbi.nlm.nih.gov/feed/
Comments and questions about NCBI resources may be sent to NCBI at: vog.hin.mln.ibcn@ofni, or by calling 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
- NCBI News, December 2008 - NCBI NewsNCBI News, December 2008 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...