NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
PubMed Interface for Mobile Devices Now Available
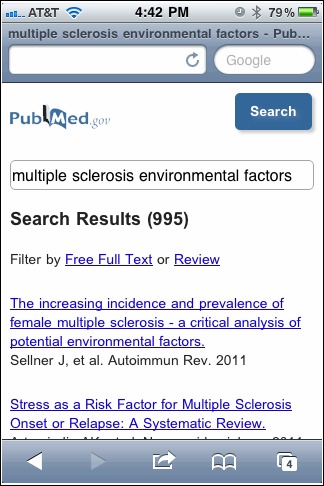
PubMed Mobile Beta is now live. This is a special lightweight web interface that makes PubMed faster to load and easier to use on smart phones and other mobile devices. The PubMed site will eventually automatically detect browsers on mobile devices and will load the new interface. For now, direct access is available at the following address.
http://www.ncbi.nlm.nih.gov/m/pubmed/
The recent PubMed Technical Bulletin provides additional information.
NCBI Bookshelf Updated to the New Entrez Design
The NCBI Bookshelf is now fully updated to the discovery-oriented Entrez design that first appeared in PubMed in 2009. Bookshelf has a new homepage, results displays, search limits, and an Advanced search page. A new browser helps to quickly find titles of interest. The NLM Technical Bulletin explains the new features in more detail.
New Organism Builds in UniGene
Two new organisms have builds in UniGene: the important plant pathogen Pythium ultimum, and the mountain pine beetle (Dendroctonus ponderosae), a serious forest pest.
Pythium ultimum (build information, 6,663 clusters, FTP) is the cause of damping-off disease in seedlings and rot in stored root vegetables. Pythium ultimum is a member of the Oomycetes, a fungi-like group of organisms that also contains the Phytophthora causative agents of potato (late) blight.
The mountain pine beetle (build information, 6,783 clusters, FTP) is a bark beetle that is a destructive pest of pine forests in the western United States. Analysis of this beetle's transcriptome should provide important insights into pheromone biosynthesis and potentially specific control mechanisms for these pests.

Root rot caused by Pythium (left); the mountain pine beetle (right).
NCBI YouTube Video Update
A new video on NCBI’s YouTube Channel shows how to display Primer-BLAST search results on the Graphical Sequence Viewer (GSV), a feature of the GSV described in the October 2011 NCBI News. This Video joins nine others that are included in the Tutorials Playlist. These videos provide brief instructions on useful features of the NCBI Web services.
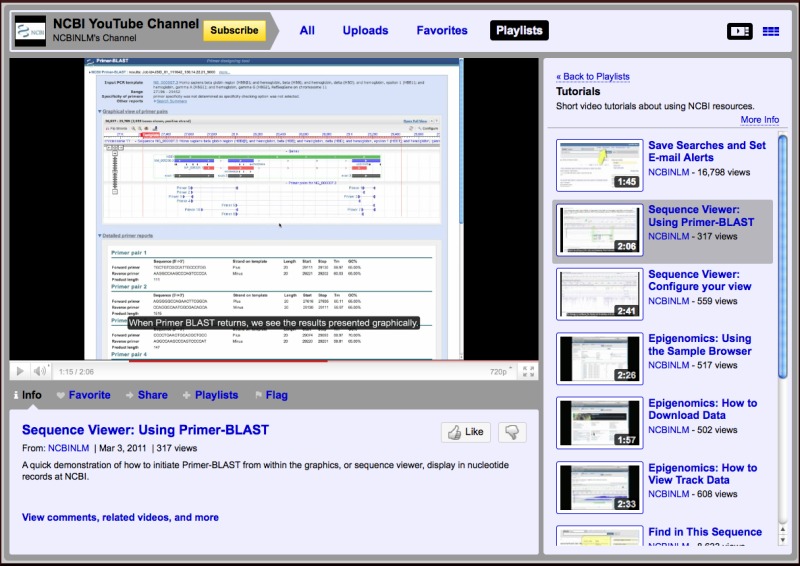
Links to relevant NCBI YouTube Videos now appear in the Discovery Column in Entrez record views or search results under certain circumstances. Specific video ads appear for only a few weeks at a time. These videos provide brief tutorials on how to use relevant features of the current page and promote new or underused but useful aspects of the service. The first of these video ad campaigns appeared in the sequence databases to introduce the Find-in-Sequence feature on sequence records described in the September 2010 NCBI News. The most recent video ad appeared on PubMed search results to promote using My NCBI to set up custom searches and e-mail alerts. The video portlet that is embedded in the page allows the video to be played in position, helpful for providing instruction within the current view. Alternatively the larger version of the video may be played on YouTube by clicking the “See larger video at YouTube” link.
Additional video ads will appear in many of the Entrez databases in the future.
RefSeq News
RefSeq Release 46 is available through Entrez, BLAST, and the RefSeq FTP site. The current release includes nearly 17 million sequence records from 11,734 organisms. The release notes provide more detailed information.
GenBank News
GenBank release 182 is available through the NCBI web and FTP sites. The current release incorporates data available as of Feb 15, 2010 and contains 124,277,818,310 bases from 132,015,054 sequence records. Release notes describe the current state of data and upcoming changes.
Microbial Genomes Update
Sixty-three finished microbial genomes were released during January and February 2011. The original sequence data files submitted to GenBank/EMBL/DDBJ are available in the Bacteria directory in the genomes area of the GenBank FTP site. Fifty-nine RefSeq provisional versions were made from a selected set of finished genomes.
In addition, 184 microbial whole genome shotgun-sequencing projects were added to GenBank during this period. The original submitted files are available in the Bacteria_DRAFT directory in the GenBank genomes area. RefSeq provisional versions of 55 of these projects are also available.
All GenBank and RefSeq microbial genomes are incorporated in the NCBI integrated Entrez search and retrieval system.
Mouse Genome Annotation Release (build 37.2) and Updated Mouse Consensus Coding Sequence (CCDS) Data
A new mouse genome annotation (v.37.2) is now available in Entrez, the Map Viewer, BLAST, and on the FTP site. This includes the reference C57BL/6 strain genome (MGSCv37) and Celera assemblies as well as alternate regional haplotype assemblies (ALT_LOCI) designated for 12 other strains. Figure 1 highlights mouse genome and the alternate locus for the beta globin region. The mouse Consensus Coding Sequences (CCDS) have also been updated to include new coding sequences that are consistently annotated by NCBI, the European Bioinformatics Institute, Wellcome Trust Sanger Institute, and the University of California, Santa Cruz. This update adds of 4,561 new CCDS records and 2,685 Genes into the mouse CCDS set. Mouse build 37.2 includes a total of 22,187 CCDS records that correspond to 19,509 GeneIDs. The statistics report has more details. The mouse CCDS update can be downloaded from the NCBI FTP site.
HomoloGene Release 65 Now Available
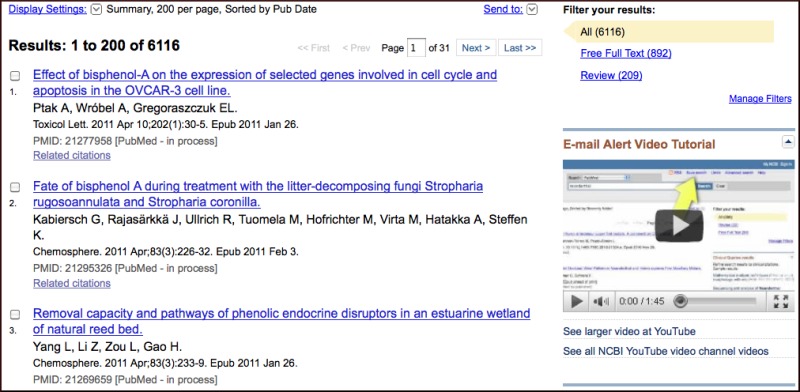
The new HomoloGene release 65 includes updated annotations for human, zebrafish, fruitfly, C. elegans, and Arabidopsis thaliana. Clusters also feature an updated related UniGene section as shown in Figure 2 that groups linked UniGene records by organism. The HomoloGene homepage has the latest statistics.

Figure
Figure 2. HomoloGene Cluster 55465 containing vertebrate CFTR homologs. Top panel, gene and sequence cluster. Center panel, new related UniGene section with organism summaries. Bottom panel, related UniGene clusters for bony fishes.
Genome Workbench Version 2.2.2 Release
Genome Workbench is NCBI’s standalone genome analysis and annotation tool. The new Genome Workbench release has several important enhancements including: integration of WindowMasker, improved displays of tree views, better network support for clients behind firewalls, improved support for international symbols, new status reporting for background work, and updated documentation. The Genome Workbench homepage has more information and links to extensive help documentation and access to download the compiled program for several versions of Windows, Mac, and Linux operating systems as well as the C++ source code for building Genome Workbench on all platforms.
NCBI Responds to a Report of Contamination in the Sequence Databases
A report by Longo et al. in the on-line journal PLoS One provides examples of cross-species contamination in DNA sequence data. The reported contamination is almost entirely in high-throughput, low-coverage sequences and is removed by filters in the NCBI genome pipeline for finished genomes. The full response is available at the NCBI website.
NCBI Discontinues the Short Read Archive, Trace Archive, and Peptidome
Due to budget constraints, NCBI has discontinued the Peptidome repository for protein/peptide mass spectroscopy data and will be discontinuing the Sequence Read Archive (SRA) and Trace Archive repositories for high-throughput sequence data. Data for these projects will continue to be available on the NCBI FTP site for the foreseeable future. The full announcement with additional details and information on the future disposition of data associated with these projects is available on the NCBI website.
- PubMed Interface for Mobile Devices Now Available
- NCBI Bookshelf Updated to the New Entrez Design
- New Organism Builds in UniGene
- NCBI YouTube Video Update
- RefSeq News
- GenBank News
- Microbial Genomes Update
- Mouse Genome Annotation Release (build 37.2) and Updated Mouse Consensus Coding Sequence (CCDS) Data
- HomoloGene Release 65 Now Available
- Genome Workbench Version 2.2.2 Release
- NCBI Responds to a Report of Contamination in the Sequence Databases
- NCBI Discontinues the Short Read Archive, Trace Archive, and Peptidome
- NCBI News, March 2011 - NCBI NewsNCBI News, March 2011 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...