NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
New Databases and Tools
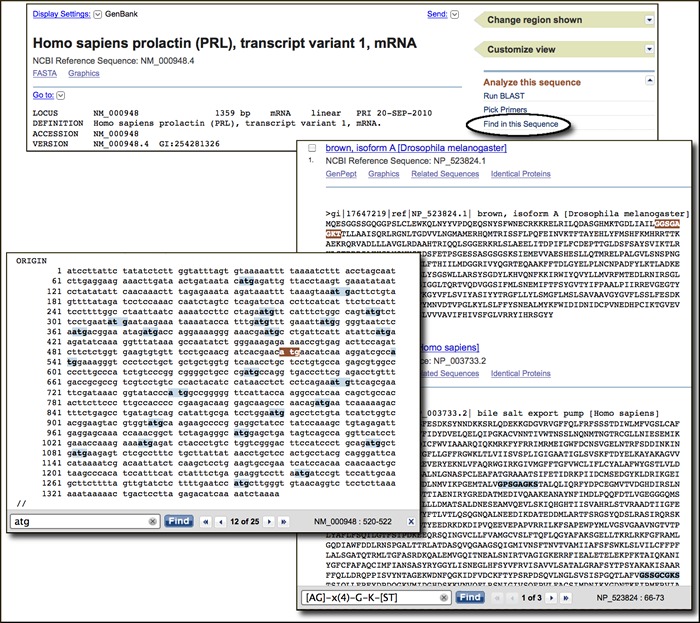
Find-in-Sequence now available in nucleotide and protein databases
The new Find-in-Sequence search finds and highlights subsequences, patterns, or motifs in nucleotide or protein sequences displayed in the Entrez system. In nucleotide sequences, sub-sequences or patterns of interest may include restriction enzyme recognition sites, transcription factor binding sites and other promoter elements, poly-adenylation signals, start and stop codons, and many others. In proteins, these may include potential cleavage locations, sites of potential post-translational modification, motifs representing active sites, cofactor binding pockets, or other structural or functional signatures. The link to open the Find-in-Sequence search box appears in the Analysis Tools portlet in the right-hand Discovery column of nucleotide and protein record views. Find-in-sequence works with single and multiple sequence displays with any format that shows the sequence (GenBank, GenPept, FASTA). The search box that opens at the bottom of the display has the familiar look and feel of the find in page function of the Web browser showing all matches and providing means to jump to the next or previous match. However, unlike the simple find function, Find-in-Sequence allows matching across spaces and line breaks in the formatted sequence and can use standard nucleotide and protein ambiguity codes as well as Prosite patterns for protein sequences. Find-in-Sequence provides rapid mapping of custom features onto nucleotide and protein sequences and is a powerful addition to the suite of analysis tools now available directly from sequence records in the Entrez system.
A video demonstrating this feature is now available on the NCBI YouTube channel.
Recent NCBI Publication on dbVar
The NCBI Database of Genomic Structural Variation (dbVar) announced in the February 2010 issue of the NCBI News was described in a recent correspondence article in Nature Genetics.
Church DM, Lappalainen I, Sneddon TP, Hinton J, Maguire M, Lopez J, Garner J, Paschall J, DiCuccio M, Yaschenko E, Scherer SW, Feuk L, Flicek P. Public data archives for genomic structural variation. Nat Genet. 42, 813-814 (2010). PMID: 20877315
The publication and resource are also announced in a recent NIH press release.
dbVAR provides information on large scale (> 1Kb) variations in genome sequences and includes copy number variants and other deletions and insertions.
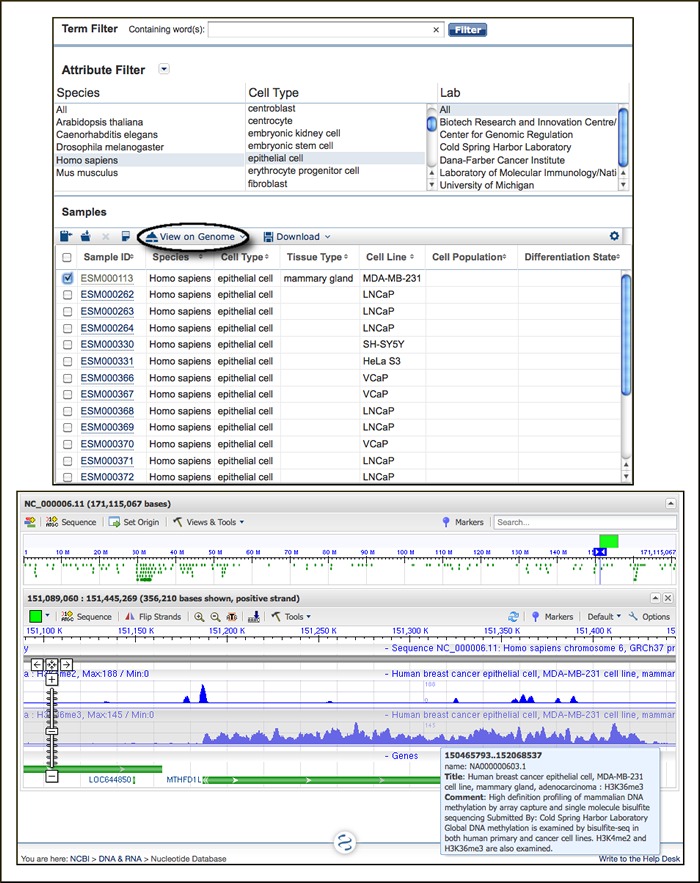
Epigenomics Resource
A new Epigenomics resource (www.ncbi.nlm.nih.gov/epigenomics) is part of the Entrez search and retrieval system. Epigenomics is a new field of research with the goal of understanding how different cell types and lineages acquire distinct patterns of gene expression. The Epigenomics database contains results of genome-wide studies on modifications of chromatin (histone modification, DNA methylation, DNAase footprinting) in various cell types. These studies assay programmable changes that affect gene expression (epigenetics). A Sample Browser provides access to experimental data. Data can be filtered based on biological attributes such as cell type, differentiation stage, health status, and others. Data may be displayed on the genome using either the UCSC genome browser or NCBI graphical sequence viewer as shown below.
Microbial Genomes
Eleven finished microbial genomes were released during September 2010. The original sequence data files submitted to GenBank/EMBL/DDBJ are available on the FTP site: ftp.ncbi.nih.gov/genbank/genomes/Bacteria/. The RefSeq provisional versions of these genomes are also available: ftp.ncbi.nih.gov/genomes/Bacteria/.
GenBank News
GenBank release 179.0 is available via web and FTP. The current release includes information available as of August 16, 2010. Release notes are available on the on the NCBI ftp site: ftp.ncbi.nih.gov/genbank/gbrel.txt
Updates and Enhancements
dbGaP
The Entrez dbGaP resource has migrated to the streamlined discovery-oriented design that has been in service in PubMed for nearly a year, described fully in the November 2009 issue of the NCBI News. Included in the re-design is a new home page (www.ncbi.nlm.nih.gov/gap), Limits and Advanced Search page. The image of the human face on the homepage, shown immediately below, is a composite of faces from approximately 10,000 people.
PubMed DTDs
PubMed E-Utility 2011 DTDs will be updated in mid-December, approximately on December 13. The new DTDs can be found:
http://eutils.ncbi.nlm.nih.gov/corehtml/query/DTD/pubmed_110101.dtd
RefSeq
RefSeq Release 43 is now available through the Entrez system and can be downloaded from the FTP site (ftp.ncbi.nlm.nih.gov/refseq/release). This full release incorporates genomic, transcript, and protein data available as of September 16. It includes 11,223,078 records from 10,854 different species and strains. Changes since the last release are desciribed in the release notes (ftp.ncbi.nlm.nih.gov/refseq/release/release-notes/RefSeq-release43.txt). More information on the RefSeq project is available on the RefSeq Homepage (www.ncbi.nlm.nih.gov/RefSeq/).
Entrez Gene Record Status
Entrez Gene (www.ncbi.nlm.nih.gov/gene) has two new properties field terms to identify the status of records that are no longer current (alive). Replaced records, retrieved with the search term “replaced”[Properties], are those that have been made secondary to (merged with) another gene record while discontinued records, retrieved with “discontinued”[Properties], are no longer current but not identified with another record. As before, current records can be retrieved with the term “alive”[properties] or by using "current only"[Filter].
NCBI Workshop at ASHG Annual Meeting
NCBI scientists will present a special workshop at the American Society for Human Genetics meeting on November 4 at 7:15 p.m. The workshop entitled, “A Practical Guide to Genome-Scale Data at NCBI”, will provide information on genome-scale resources available at NCBI including finding and downloading data, analysis, and management of data sets. NCBI will also staff an exhibit booth at the meeting.
Announce Lists and RSS Feeds
Eighteen topic-specific mailing lists are available which provide email announcements about changes and updates to NCBI resources including dbGaP, BLAST, GenBank, and Sequin. The various lists are described on the Announcement List summary page: www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html. To receive updates on the NCBI News, please see: www.ncbi.nlm.nih.gov/About/news/announce_submit.html.
Twelve RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce. Please see: www.ncbi.nlm.nih.gov/feed/.
Users can also stay updated on NCBI’s resources on Facebook and Twitter: twitter.com/NCBI.
Send comments and questions about NCBI resources to: vog.hin.mln.ibcn@ofni, or by calling 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
- NCBI News, September 2010 - NCBI NewsNCBI News, September 2010 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...
