NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997.
The means by which the viral glycoproteins are included as part of the emerging virion, largely to the exclusion of cellular proteins, is not understood. It is generally thought that Gag proteins may play a part in this process, even though the Env glycoproteins themselves are not needed for budding. The portion of Gag most likely to be involved in Env packaging is the MA sequence, which is closely associated with, and perhaps partially buried within, the virion membrane close to TM (Pepinsky and Vogt 1979, 1984; Pepinsky 1983). Indeed, it has been shown that the MA protein of ASLV can be chemically crosslinked to TM protein in the mature virus (Gebhardt et al. 1984), and ultrastructural examinations of HIV-1 particles have revealed a correlation in the distribution of Gag and Env (Bugelski et al. 1995). Data supporting two different models of Env packaging are discussed below.
Passive Packaging
There is much evidence that Gag-Env interactions do not have an obligatory role in assembly. In particular, it has been shown that the short cytoplasmic tail of the ASLV Env protein can be deleted without loss of packaging or infectivity (Perez et al. 1987). Moreover, a recombinant of ASLV in which the env gene is replaced with the coding sequence for influenza virus hemagglutinin (HA) makes particles that contain this foreign glycoprotein and are infectious (Dong et al. 1992a). Because the sequence of HA, including its TM and cytoplasmic domains, is unrelated to Env, it seems rather unlikely that one could replace the other if a specific interaction with Gag was required. This capacity for packaging foreign glycoproteins (pseudotyping) is not restricted to ASLV but appears to be a property of all retroviruses (Chapter 3. Moreover, HIV-1 and SIV also have the ability to package their glycoproteins even after significant truncation of the cytoplasmic tail of their TM proteins (Johnston et al. 1993; Freed and Martin 1995, 1996; Mammano et al. 1995; see below).
Further evidence against obligatory Gag-Env interactions has been gathered from studies in which the human CD4 molecule was expressed in RSV-infected avian cells. This unrelated foreign glycoprotein was found to be packaged on viral particles along with the Env glycoproteins (Young et al. 1990). The amount of CD4 associated with the particles was quite high and was readily detected biochemically. Other (avian) glycoproteins were excluded from the particles, and thus the packaging of CD4 would seem to be a specific event.
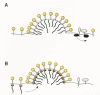
To explain the results obtained with RSV, a passive model for Env glycoprotein packaging (Fig. 12A) has been proposed (Young et al. 1990; Hunter 1994), which is based on the fact that host proteins on the surface of a cell are there for a purpose (e.g., signal transduction) and participate in interactions with other cellular proteins. Because of this, their movement within the plasma membrane is restricted, which thereby prevents them from being packaged into the viral particles that emerge from the cell surface as Gag proteins push through. In contrast, viral glycoproteins such as Env and HA do not interact with other cellular proteins. This allows them to flow into the sites of budding, where they can be passively incorporated into particles. In this model, human CD4 is available for packaging in avian cells because it is not anchored to the cellular proteins with which it normally interacts (e.g., the Lck protein on the inner surface of human lymphocytes; see Ravichandran et al. 1996).
Active packaging
Studies of HIV-1 have provided evidence for a specific interaction between Gag and Env proteins (Fig. 12B). With regard to Gag, many deletions and substitutions in the MA sequence have been found that abolish glycoprotein packaging, even though budding and particle release continue (for a summary, see Massiah et al. 1994; see also Yu et al. 1992; Wang et al. 1993; Lee and Linial 1994; Freed and Martin 1995, 1996). Moreover, deletions of the cytoplasmic domain of the TM protein have been found that also affect glycoprotein packaging (Dubay et al. 1992b; Yu et al. 1993; Brody et al. 1994b; Freed and Martin 1996). More recently, a gain-of-function experiment was reported that further supports the idea of an MA-Env interaction (Dorfman et al. 1994a). Specifically, a chimera was made in which the MA sequence of the visna virus Gag protein was replaced with the MA sequence from HIV-1. This Gag chimera produced particles that were capable of packaging the glycoproteins of HIV-1 but not those of visna virus. Interaction between HIV-1 Gag and Env stabilizes the Env protein on the surface of the cell where it is otherwise rapidly internalized (Egan et al. 1996; Lee et al. 1997).
MA mutants of HIV-1 that are defective for packaging wild-type Env proteins nevertheless retain the capacity for packaging Env glycoproteins with short cytoplasmic tails, whether these are from other retroviruses (e.g., MLV) or are versions of HIV-1 Env from which the tail has been deleted (Freed and Martin 1995, 1996; Mammano et al. 1995). On the basis of these results, retroviruses with tails that are normally short would not be expected to exhibit the exclusion of their Env proteins as a result of MA mutations. Consistent with this, all MA deletion mutants of the ASLV and MLV Gag proteins that retain the ability to make particles also retain infectivity and hence the capacity to package their glycoproteins (JØrgensen et al. 1992; Granowitz and Goff 1994; Nelle and Wills 1996).
Further evidence for an interaction between Gag and Env proteins comes from studies of HIV-1 budding in polarized epithelial cells. When such cells become confluent in culture, tight junctions form and the monolayer exhibits polarity (Matter and Mellman 1994). The top of the monolayer (toward the growth medium) is equivalent to the apical surface of actual epithelial cells, which faces the lumen of organs such as the intestine; the bottom is equivalent to the basolateral membrane, which faces the circulatory system. In polarized cells, enveloped viruses bud from the apical or basolateral cell membrane, depending on the virus (Tucker and Compans 1993). Retroviruses bud baso-laterally (Roth et al. 1983; Owens and Compans 1989), whereas influenza virus, for example, buds apically.
It has been long known that viral glycoproteins are sorted to the appropriate cell surface when expressed in polarized cells in the absence of other viral components, and this suggests that they may have a critical role in determining the site of assembly (Tucker and Compans 1993). However, unlike other enveloped viruses, retroviruses do not require their glycoproteins to be present for budding to take place, and in polarized cells, the HIV-1 Gag protein directs budding from both cell membranes. Nevertheless, when the HIV-1 Env proteins are coexpressed with Gag, budding is observed only from the basolateral membrane (Owens et al. 1991; Lodge et al. 1994). This suggests that an overriding (dominant) Gag-Env interaction takes place. The TM protein (gp41) is responsible for redirecting Gag to the basolateral membrane, and deletions in its cytoplasmic tail abolish the effect. Moreover, MA deletion mutants that are incapable of packaging Env fail to exhibit polarized budding, even when the wild-type Env proteins are coexpressed. Interestingly, HIV-1 has been found to bud in a polar manner even in infected lymphoid cells and only at sites where Env antigens colocalize (Bugelski et al. 1995), again suggesting that Gag proteins “home” to regions of the plasma membrane where the glycoproteins reside.
- Incorporation of Env into the Viral Particle - RetrovirusesIncorporation of Env into the Viral Particle - Retroviruses
- Bacillus subtilis strain DF60 16S ribosomal RNA gene, partial sequenceBacillus subtilis strain DF60 16S ribosomal RNA gene, partial sequencegi|42565626|gb|AY462216.1|Nucleotide
Your browsing activity is empty.
Activity recording is turned off.
See more...