NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997.
To initiate the infection cycle, all viruses require an interaction between a surface molecule—the receptor—and a protein or proteins on the surface of the virion. This interaction is highly specific. Small molecular changes at the binding site of the receptor can render a cell virtually uninfectable by the cognate retrovirus, in that the infection efficiency can drop by six orders of magnitude or more. Receptor specificity is of considerable importance to the biology of the virus: The distribution of suitable receptors among animal species determines the host range of the virus, and the distribution of receptor expression among cells in the body is the first (but not the only) determinant that specifies the pathogenic outcome of infection. For these reasons, the molecular identification of receptors is an important, and often difficult, step in understanding virus-host interactions.
The identity of the host-cell-encoded receptor molecule(s) that allows attachment and entry of retroviruses was until quite recently unknown for any retrovirus. The initial experimental approaches involved attempts at identifying host-cell molecules that bound to soluble forms of the SU protein (DeLarco and Todaro 1976; Moldow et al. 1979; Landen and Fox 1980; Robinson et al. 1980; Choppin et al. 1981). However, the results of these studies were inconsistent and no direct role in viral entry could be assigned. Other direct approaches that attempted to identify receptor proteins, such as anti-idiotype antibodies (Davis et al. 1992), were no more successful.
Instead, successful identification of retroviral receptors was both motivated and abetted by a special property. Within most animal virus families, there is little or no variation in receptor usage among closely related viruses (those classified in the same genus). Several retroviral groups, however, are unusual in this respect. The avian and mammalian C-type viral genera are both characterized by considerable polymorphism in receptor expression in the host species and receptor usage by the individual viruses within each genus. Different members of the same host species can differ in the presence or absence of functional receptors for specific viral subgroups, and closely related viruses can use completely different receptors.
The receptors used by different viruses can be further identified by the useful property of interference, or superinfection resistance. Since cells infected by retroviruses often survive and produce virus, infected cells can be tested for their susceptibility to the same or different viruses. Although such infected cells are readily superinfected by viruses that use different receptors, if the same receptor is used by both viruses, then the Env protein of the virus being produced can interact with the receptor and prevent superinfection by the second virus (for review, see Weiss 1993). This effect permits viruses to be readily assigned to subgroups as defined by receptor usage.
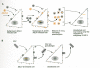
Another useful technique is the generation of pseudotypes, in which coinfection of cells with two retroviruses, or one retrovirus and the unrelated rhabdovirus vesicular stomatitis virus (VSV), leads to the production of viral particles that have incorporated functional Env proteins from one virus into a virion derived from an unrelated genome. This approach allows the identification of receptors on the surface of cells that cannot otherwise be infected by the virus of interest (Fig. 1) (Boettiger 1979; Weiss 1980).
The genetic differences in receptor distribution and usage, although they can create rather confusing biological data, underlie the most incisive cloning approaches to receptor isolation. With the exception of HIV/SIV, all known retroviral receptors were cloned by a strategy that relies on the identification of DNA sequences capable of rendering nonpermissive cells susceptible to infection. The genetics of receptor function can then be used to confirm the identity of the cloned sequence and to identify the specific amino acid residues with which the virus interacts. Finally, the genetics of the virus can be used to probe the molecular interaction between the receptor and the virus from a viral perspective, as described in the next section. Because there are major differences in the nature of the receptor and its genetics, structure, and normal function from one viral group to another, this section will treat each viral group as a separate example.
Avian Sarcoma/Leukosis Viruses
A detailed analysis of viral infectivity on cells derived from different chicken embryos led to the discovery of a distinct pattern of susceptibility and resistance to infection (for review, see Vogt and Hu 1977). No less than nine subgroups of these viruses, named A through J, representing distinct patterns of receptor usage have been identified (Table 1). The best studied viruses isolated from chickens can be divided by interference patterns and other properties into five subgroups, A through E (Table 2).
Table 1
Subgroup/Receptor Classification of ASLV.
Table 2
Receptor Interference among ASLV Strains.
Genetics of ASLV Receptors
The patterns of viral infectivity are consistent with three different genetic loci, termed tv-a, tv-b, and tv-c, encoding determinants of susceptibility to different viruses (Rubin 1965; Vogt and Ishizaki 1965; Payne and Biggs 1966). tv-a and tv-c control susceptibility to subgroup A and C viruses in a straightforward way, whereas tv-b is responsible for infection by B, D, and E viruses. Subgroup B and D viruses use the same receptor on chicken cells but differ in the greater ability of D subgroup viruses to infect mammalian cells.
The phenotype of chicken cells is defined by their resistance to infection by a specific subgroup virus, rather than by their susceptibility. For example, C/A (“C bar A”) indicates chicken cells resistant to subgroup A viruses, and C/O indicates susceptibility to all viral subgroups. The most commonly used birds have a C/E phenotype. Resistance in this system is essentially absolute, in that a viral stock with an titer of greater than 1 × 106 infectious units per milliliter will be incapable of infecting resistant cells.
The results of the genetic studies are interpreted to mean that three chicken loci encode receptors for the viral subgroups A through E, with resistance to infection indicating the absence of a specific functional receptor. In this and all other cases, it is important to realize that the absence of a functional receptor does not mean the absence of the receptor protein per se (Weiss 1993). Rather, as is more clearly demonstrated with the ecotropic MLV receptor, single amino acid changes can completely abrogate receptor function without affecting the normal function of the protein. This principle is likely to apply in the ASLV case, but it has not been demonstrated to date.
The ASLV Subgroup A Receptor Is an LDLR-related Protein and Is Unrelated to the Subgroup B Receptor Transfection of chromosomal DNA from susceptible chickens into resistant mouse 3T3 cells allowed the cloning of an 8-kb fragment of genomic DNA that could transfer susceptibility to subgroup A ASLV (ASLA-A) (Young et al. 1993). Surprisingly, neither transcripts nor a protein product of this locus could be detected in chicken cells, although its identity with tv-a was confirmed by the close genetic linkage of sequences hybridizing to the cloned gene and susceptibility to subgroup A virus (Bates et al. 1993).
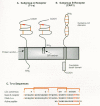
To identify the functional coding sequences of the putative receptor gene, a smaller (5.5 kb) homologous chromosomal DNA fragment, which could efficiently transfer susceptibility, was isolated from quail cells and introduced into an MLV vector as an exon-trapping approach (see Chapter 9) (Bates et al. 1993). Two remarkably small cDNA clones derived from alternately spliced transcripts of 819 and 968 nucleotides were isolated that could confer susceptibility. These cDNAs potentially code for membrane-anchored glycosylated proteins of 101 and 138 amino acids, respectively. The amino-terminal 92 amino acids of both proteins are identical, encoding a putative 83-amino-acid extracellular domain. The two proteins differ in their mode of association with the cell membrane (Fig. 2A), implying that the nature of this association is not important for receptor function. A soluble secreted form of the extracellular domain blocks subgroup A viral infection of avian cells by binding specifically to ASLV-A Env (Connolly et al. 1994; Gilbert et al. 1994). Antiserum to the product of the 819-bp transcript precipitates the product of the cloned receptor cDNA expressed in mouse 3T3 cells and specifically inhibits subgroup A viral infection of both receptor-expressing 3T3 cells and susceptible avian cells. The failure of this antiserum to precipitate detectable amounts of protein from avian cells (Bates et al. 1993) implies that the native receptor must be expressed at very low levels, and its identity remains to be determined.
The normal function of the tv-a-encoded receptor also remains to be determined. The putative extracellular domain of the cloned receptor contains a sequence closely related to the ligand-binding repeat of the low-density lipoprotein receptor (LDLR) (Fig. 2C). This repeat, which has six cysteine residues spaced over an approximately 40-amino-acid region and is reiterated seven times in LDLR, is also found as a single copy in several terminal components of the complement pathway and has been postulated to be important for extracellular protein-protein interactions (Stanley 1989). Mutagenesis of the receptor gene has indicated that the LDLR repeat region alone can function as an ASLV-A receptor if it is fused to a membrane anchor and that only a few residues within the carboxyl terminus of the repeat, which are unique to the quail and chicken receptors, have a key role in binding of the viral glycoprotein (Fig. 2C). In fact, only three amino acids—Asp-46, Glu-47, and Trp-48 between the fifth and sixth cysteines and a putative disulfide bond between the fourth and sixth cysteines—are important for receptor activity, suggesting that they interact directly with the Env protein. Moreover, a small, 19-amino-acid peptide containing these determinants specifically inhibits subgroup A viral infection (Bélanger et al. 1995; Zingler et al. 1995).
Given the overall homology between the glycoproteins of different ASLV subgroups (see below), it seemed plausible that receptors for these viruses might also be members of an LDLR-related group. However, a cellular receptor (presumably tv-b) for subgroups B and D ASLV was recently cloned using the same strategy and was identified as a rather different kind of cell surface protein, resembling a receptor for certain cytokines such as tumor necrosis factor (TNF) (Fig. 2B) (Brojatsch et al. 1996). These subgroups of ASLV have long been known to induce significant cytopathic effects in infected cells (Weller and Temin 1981), an effect that is specific for viruses that use the subgroup B receptor (Dorner and Coffin 1986). An interesting feature of this receptor is the presence of a cytoplasmic region similar to the defined “death domain” of the type I TNF receptor and Fas, which transmit signals for apoptosis (programmed cell death). In fact, quail QT6 cells expressing this protein are sensitive to apoptosis induced by a subgroup B ASLV SU-Ig fusion protein, indicating that cytopathic ASLV Env-receptor interactions might contribute to the cell killing associated with subgroup B and D viral infections (Brojatsch et al. 1996).
Mammalian Type-C Retroviruses
The MLVs have been classified into four different host-range subgroups according to the distribution of their distinct cell surface receptors among species and according to their viral interference patterns (Table 3). The receptor for ecotropic viruses is restricted to cells of mouse or rat origin, whereas the receptor for viruses with a xenotropic host range is not found on mouse cells but is present on cells of a variety of other species. The other two subgroups of viruses, the amphotropic and dual- or polytropic viruses, use receptors found on both rodent cells and cells of other species, but they do not interfere with one another, indicating that the receptors are distinct molecules.
Table 3
Receptor Interference between Murine Type-C Viruses (MLV) in Murine Fibroblasts.
Genetics of MLV Receptors
The ecotropic viruses were the first class of MLVs to be identified. They comprise some of the most intensely studied retroviruses, including exogenous viruses such as Moloney MLV (Mo-MLV) and Friend MLV (Fr-MLV), as well as the endogenous virus of the AKR mouse strain, AKR MLV (AKV). A functional receptor for the ecotropic MLVs is encoded by all strains of laboratory and wild mice of the genus Mus musculus that have been examined to date. The assignment of a single genetic locus for susceptibility to ecotropic MLV infection was consistent with a single gene encoding the receptor protein. The evidence for this came from studies of somatic cell hybrids that were created by fusion of susceptible mouse cells to nonpermissive hamster cells. When cells that retained susceptibility to ecotropic MLV infection were characterized for their mouse chromosomal content, it was possible to assign the receptor gene(s) to the Rec1 locus on chromosome 5 (Gazdar et al. 1977; Oie et al. 1978; Ruddle et al. 1978).
Xenotropic and polytropic MLVs are present as endogenous proviruses in all inbred mice (see Chapter 8). Like ASLV receptors in chickens, the receptor for xenotropic MLV is polymorphic in mice but not in other species. Although xenotropic viruses will not infect cells from inbred strains of mice, they will infect cells derived from some wild mice and species of mice other than M. musculus (Hartley and Rowe 1975; Lander and Chattopadhyay 1984). The genetic locus for susceptibility maps to chromosome 1, and it is inseparable from the gene for the polytropic viral receptor, suggesting that alleles of this gene can serve as the receptor for both polytropic and xenotropic viruses (Kozak 1985; Hunter et al. 1991)
Xenotropism—the inability of an endogenous retrovirus to infect cells of the species whose germ line it inhabits—is not limited to the endogenous viruses of mice but is also seen with the subgroup E viruses of chickens and with endogenous viruses of cats and primates. It is likely that xenotropic viruses originally inserted into the germ line in a host background that encoded their cognate receptor but that the functional xenotropic viral receptor allele was subsequently lost, probably under selective pressure from exogenous xenotropic viruses.
Amphotropic viruses are exogenous MLVs originally isolated from some wild mouse strains. Because of their efficient infection of human cells, the amphotropic env gene is widely used in retroviral vectors (Chapter 9). The distribution of receptor activity for amphotropic viruses among species differs from that of xenotropic and polytropic viruses, as does the map location of the receptor (Garcia et al. 1991). To date, genes encoding receptors for ecotropic and amphotropic MLVs have been cloned and characterized; those for xenotropic and polytropic viruses remain to be discovered.
The Ecotropic MLV Receptor, a Cationic Amino Acid Transporter
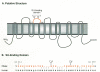
As with the ASLV receptors, the cloning and identification of a gene for the ecotropic MLV receptor (Albritton et al. 1989) used the transfer of mouse chromosomal DNA into resistant human cells to search for sequences that could confer susceptibility to ecotropic viral infection. This approach identified a novel gene that encodes a membrane protein of 622 amino acids (molecular weight of 67,000) and which appears to contain as many as 14 membrane-spanning domains (Fig. 3A). Expression of the gene in human cells not only confers susceptibility to ecotropic viral infection, but also results in an eightfold increase in binding of soluble ecotropic glycoprotein (gp70) to the transfected cells (Albritton et al. 1993). Consistent with the mapping of Rec1 in mouse-hamster hybrid cell studies, the receptor gene mapped to mouse chromosome 5 (Table 4) (Kozak et al. 1990).
Table 4
Receptors for Mammalian Type-C Retroviruses.
Localization of possible binding sites for the MLV Env protein has been aided by identification of a highly related (87% identity) gene in human cells that maps to chromosome 13 (Yoshimoto et al. 1991; Albritton et al. 1992). Exchanging portions of the genes that encode the permissive mouse and nonpermissive human molecules allowed identification of amino acid residues in the third extracellular loop, where significant differences in sequence exist, required for both gp70 binding and MLV infection (Fig. 3B) (Albritton et al. 1993; Yoshimoto et al. 1993). Surprisingly, certain substitutions of mouse residues into the human sequence confer susceptibility to infection without causing the enhanced levels of gp70 binding seen with the wild-type murine protein (Albritton et al. 1993). Thus, it is likely that MLV receptors can vary significantly in their affinities for the viral glycoprotein while still retaining the ability to mediate viral entry. This property is similar to that described for the HIV gp120 and its receptor CD4. Variations within the sequence of the third extracellular loop of the ecotropic MLV receptor can result in loss of receptor function for one ecotropic virus, whereas susceptibility to others is maintained. This is the case for the receptor from the feral mouse, Mus dunni, in which a single isoleucine to valine substitutionprevents entry of Mo-MLV without affecting the Rauscher or Friend strains of ecotropic virus (Eiden et al. 1993). In hamster cells, the ecotropic glycoprotein-binding site appears to be masked or modified by the presence of an oligosaccharide side chain since treatment of Chinese hamster ovary (CHO) or BHK21 cells with the glycosylation inhibitor tunicamycin renders the cells sensitive to infection (Wilson and Eiden 1991; Miller and Miller 1992).
The potential for the ecotropic receptor molecule to span the membrane multiple times suggested at the outset that it might function in the cell as a membrane transporter protein (Albritton et al. 1989). Kim et al. (1991) also noted a coincidence in the positions of the first eight putative membrane-spanning domains of this receptor and those of the arginine and histidine permeases of the yeast, Saccharomyces cerevisiae. This relationship was confirmed when it was shown that injection of Xenopus oocytes with the messenger RNA encoding the ecotropic MLV receptor results in an increased, stereospecific, saturable, and sodium-independent uptake of L-arginine, L-lysine, and L-ornithine (Kim et al. 1991; Wang et al. 1991). The characteristics of the permease, now termed mCAT-1, encoded by the ecotropic viral receptor gene are indistinguishable from that of y+, the previously characterized, principal transporter of cationic amino acids in mammalian cells. In addition, the distribution of mCAT-1 mRNA expression in different tissues is consistent with that described for y+ (Kim et al. 1991). The gene for mCAT-1 is now known as Atrc1 (for amino acid transporter, cationic 1) (see Table 4).
The amino acid transporter property of mCAT-1 can also be dissociated from its function as a receptor, since mutations that destroy transporter activity do not prevent the molecules from functioning efficiently as receptors (Wang et al. 1994). Conversely, binding of MLV Env protein only partially inhibits transporter activity (see below). Moreover, chimeras containing the first three extracellular loops of mCAT-1 and the carboxy-terminal portion of a second murine cationic amino acid transporter, mCAT-2, function as receptors for ecotropic MLV but have transporter activities characteristic of the mCAT-2 donor (Closs et al. 1993; Kavanaugh et al. 1994b).
The Receptors for GALV, FeLV-B, and Amphotropic MLV Are Phosphate Symporters
The receptor for GALV
The host range of GALV is extremely broad and includes cells of human, monkey, cat, dog, cow, bat, mink, rabbit, and rat (but not mouse) as well as a variety of avian species (chicken being the exception) (Teich 1984). Since all of the isolates belong to a single interference group (Rangan 1974), it appears that the receptor for GALV is widely conserved across diverse species.
Using an experimental strategy similar to that employed for identification of the ecotropic MLV receptor, O'Hara et al. (1990) identified and cloned a human gene that could confer susceptibility to GALV infection to mouse fibroblasts that were previously resistant to this virus. The presumptive protein encoded by the gene, originally called GLVR1, is 679 residues in length with ten potential membrane-spanning domains (Fig. 4A). The gene was mapped to human chromosome 2 by in situ hybridization and somatic cell hybrid analysis, and the murine homolog, Glvr1, was similarly localized to a region of mouse chromosome 2 (Table 4) (Adamson et al. 1991; Kaelbling et al. 1991). The sequence of the murine protein differs from that of the human receptor molecule in 10% of its residues. By constructing chimeric and mutant proteins, it was possible to map a region important for receptor function; amino acids 550 and 551, within a domain that is highly polymorphic between species (Fig. 4B), are critical for GALV entry (Johann et al. 1992, 1993).
The sequence of GLVR1 implies homology with that of the Neurospora phosphate permease gene, pho-4+, the amino- and carboxy-terminal thirds of the molecules showing 31% and 38% identity, respectively (Johann et al. 1992). In both proteins, there is an internal repeat corresponding approximately to the amino- and carboxy-terminal regions of identity, which may have resulted from an ancient duplication. GLVR1 differs most dramatically from the phosphate permease in its possession of a long, potentially cytoplasmic, loop of more than 250 amino acids between membrane-spanning domains 6 and 7 (Fig. 4A). The presence of multiple membrane-spanning domains, as well as the homology with pho-4+, provided strong evidence for a membrane transporter role for GLVR1. Kavanaugh et al. (1994a) confirmed that this protein is indeed a sodium-dependent phosphate symporter. It is therefore appropriate to rename this receptor hPiT-1 (for human inorganic phosphate transporter). The human gene has been designated SLC20A1 (for solute carrier).
GALV and FeLV-B Share a Common Receptor
Sommerfelt and Weiss (1990) observed cross-interference between GALV and subgroup B FeLV, suggesting that these viruses might share a common receptor. Additional evidence for this supposition came from the observation that mouse fibroblasts expressing hPiT-1 are susceptible to FeLV-B infection (Takeuchi et al. 1992). Moreover, using murine retroviral vectors pseudotyped with either the GALV or FeLV-B glycoproteins, it was possible to demonstrate reciprocal interference on GALV-infected and FeLV-B-infected cells. Amino acid changes introduced into the highly polymorphic domain that spans residues 550–558 block FeLV-B and simian sarcoma-associated virus (SSAV) infection of cells without affecting the receptor function for GALV (Tailor et al. 1993), a situation reminiscent of the M. dunni mCAT-1 protein's inability to function as a receptor for Mo-MLV (Eiden et al. 1993).
The Amphotropic MLV Receptor
The amphotropic MLV receptor has been identified through two independent experimental approaches. In the first, Miller et al. (1994) employed transfection of a rat cDNA expression library into resistant CHO cells, followed by screening for susceptibility to infection by an amphotropic MLV vector, to identify what they have referred to as the rat Ram1 gene. In the second approach, van Zeijl et al. (1994) utilized low-stringency hybridization with a cDNA encoding hPiT-1 to isolate related phage clones from human HL-60 and placental cDNA libraries. One of these clones, pGLVR2-1, encoded a 652-amino-acid protein with 62% overall identity to hPiT-1 which could confer on hamster cells susceptibility to infection by amphotropic MLV vectors. The rat and human receptors are more than 90% identical and, like hPiT-1, contain an internal, divergent repeat of sequences 1–165 at positions 483–652. By screening a series of mouse-human somatic cell hybrids, the Ram1/GLVR-2 gene was mapped to human chromosome 8 (Table 3), consistent with previous mapping studies of the position of the Ram1 locus that were based on susceptibility to amphotropic MLV infection (Garcia et al. 1991). Ram1/GLVR-2 and hPiT-1 are highly similar (81%) in their carboxy-terminal hydrophobic domains (residues 483–652). However, a cluster of amino acid differences within the fourth extracellular loop of the amphotropic receptor has a critical role in infection by GALV. These changes appear to be the reason Ram1/GLVR-2 cannot function as a receptor for GALV (see Miller and Miller 1994; van Zeijl et al. 1994). Substitution of the carboxy-terminal hydrophobic domain of Ram1/GLVR-2 with the homologous region from hPiT-1 results in a chimeric receptor that can function to mediate entry of both GALV and amphotropic MLV (Miller and Miller 1994; Pedersen et al. 1995).
As might be expected from the homology between hPiT-1 and Ram1/GLVR-2, the latter also functions as a sodium-dependent phosphate symporter (Kavanaugh et al. 1994a). Phosphate uptake was reduced by more than 50% in mouse cells expressing the amphotropic MLV envelope glycoprotein, which binds to Ram1/ GLVR-2. Thus, this receptor molecule, whose synthesis is regulated by phosphate levels, is a major phosphate transporter (mPiT-2) in these cells (Kavanaugh et al. 1994a).
Hamster cells appear to encode amphotropic receptors that differ from those of both rat and human cells. CHO cells are normally resistant to amphotropic MLV infection but, following treatment with the glycosylation inhibitor tunicamycin, become susceptible to infection (albeit ∼1000-fold less efficiently than mouse 3T3 cells). Wilson et al. (1994) have cloned and characterized a unique form of the amphotropic receptor homolog in E36 hamster cells and have shown that it can function as a receptor for both amphotropic MLV and GALV, analogous to the synthetic, chimeric dual-tropic receptor described previously. Since these cells also encode a functional hPiT-1, this finding explains why viral interference observed after infection with GALV and amphotropic MLV is nonreciprocal (Eglitis et al. 1993). A soluble factor that can block the hamster-encoded receptor is released from the CHO cells and inhibits amphotropic MLV (and to a limited extent GALV) entry. This inhibitory factor is specific for the hamster receptor since it cannot block entry mediated by the human amphotropic receptor expressed in the same cells (or hPiT-1 expressed on human cells) (Miller and Miller 1992). The presence on CHO cells of a similar dual-tropic receptor that can be blocked by a soluble factor would explain why GALV infection is only partially inhibited in the presence of the factor.
Primate Lentiviruses: HIV-1, HIV-2, and SIV
Identification and Characterization of the HIV Receptor: CD4
Shortly after the isolation of HIV from acquired immunodeficiency syndrome (AIDS) patients, its receptor was found to be the CD4 (also known as T4) protein. This 60-kD molecule, originally described as a cell surface marker for the helper/inducer subset of mature T lymphocytes, plays an important part in immune recognition and is involved in the process of T-cell activation itself (for review, see Sweet et al. 1991). It is also expressed on a variety of cell types besides T cells, particularly macrophages, monocytes, and other phagocytic cells.
A major feature of AIDS recognized very early in the history of this disease is a selective depletion of the CD4-bearing helper/inducer population of lymphocytes (Gottlieb et al. 1981) (Chapter 11). This hallmark of AIDS correlates with a selective tropism of HIV for infection of the same subset of CD4+ lymphocytes in vitro (Klatzmann et al. 1984). The dependence of HIV on the CD4 protein for infection was first demonstrated by showing that some (but not all) monoclonal antibodies (MAbs) to CD4 could block HIV infectivity in a variety of in vitro assays. Dalgleish et al. (1984) showed that these monoclonal antibodies could prevent both the formation of syncytia that result from mixing HIV-infected cells with CD4+ uninfected cells and the infection of cells with VSV pseudotypes carrying the HIV envelope protein. Similarly, monoclonal antibodies to CD4 block the infection of peripheral blood lymphocytes by HIV (Klatzmann et al. 1984) and inhibit the binding of fluoresceinated HIV or recombinant gp120 to CD4+ cells (McDougal et al. 1985; Lasky et al. 1987).
The binding of HIV-1 to CD4+ cells involves the formation of a stable complex between the gp120 (SU) molecule of the virus and CD4; immunoprecipitation with antiserum to either molecule coprecipitates the other (McDougal et al. 1986a). Measurements of the affinity constant for this reaction have shown that gp120 and CD4 form a high-affinity complex with a dissociation constant of approximately 4 × 10–9 M per liter (Lasky et al. 1987). Direct evidence that CD4 is the receptor for HIV came from the work of Maddon et al. (1986), who demonstrated that transfection and expression of the cDNA for human CD4 in HeLa cells render these normally resistant cells permissive for HIV infection. The CD4 molecule acts as a receptor for all members of the primate lentivirus group: HIV-1, HIV-2, and a diverse group of SIV strains (Hoxie et al. 1988b; Sattentau et al. 1988). However, not all cells expressing CD4 are susceptible to infection by these viruses (see below). Although interference studies confirm a common receptor for different HIV/SIVs (Hoxie et al. 1988b; Hart and Cloyd 1990; Le Guern and Levy 1992), interference between HIV-1 and HIV-2 may be nonreciprocal (see below; Hoxie et al. 1991; Le Guern and Levy 1992).
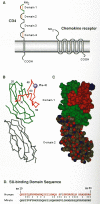
The CD4 molecule is a member of the immunoglobulin superfamily, and it consists of an extracellular region containing four immunoglobulinlike domains (D1–D4), a membrane-spanning region, and a charged cytoplasmic domain (Fig. 5) (Maddon et al. 1985). The region of CD4 that binds to gp120 was initially mapped indirectly with monoclonal antibodies to CD4 (McDougal et al. 1986b; Sattentau et al. 1986) and then through analysis of nonhuman primate cells susceptible to infection with HIV-1 (McClure et al. 1987). Subsequent immunological and molecular analyses confirmed that HIV binds to the first immunoglobulin domain of CD4; genetic analyses of gp120/CD4 binding suggested that the molecular interactions center around amino acid residues 40–60 within this region (Clayton et al. 1988; Jameson et al. 1988; Landau et al. 1988; Peterson and Seed 1988; Arthos et al. 1989; Ashkenazi et al. 1990; Bowman et al. 1990; Brodsky et al. 1990; Piatier-Tonneau et al. 1991). Moreover, substitution of residues 33–62 of human CD4 into the rat homolog renders it capable of binding gp120 and able to mediate infection of human cells (Schockmel et al. 1992; Simon et al. 1993). The three-dimensional crystal structure of the D1/D2 region of human CD4 has provided additional insight into its gp120-binding function (Ryu et al. 1990; Wang et al. 1990). Residues within the CDR2-like region of D1, shown previously by genetic and immunological approaches to be important for receptor function, are located on an exposed loop (Fig. 5) that has been postulated to interact with a cleft in gp120 (Wang et al. 1990).
Although internalization of surface CD4 molecules is observed after exposure of CD4+ T cells to phorbol esters or appropriate antigen-bearing target cells (Acres et al. 1986; Hoxie et al. 1988a), this property is not necessary for HIV entry. Initial experiments by Maddon et al. (1986) indicated that following CD4 binding, HIV entry into the cell involved endocytosis. However, it was later demonstrated that HIV enters cells through a pH-independent mechanism (Stein et al. 1987; McClure et al. 1988) and that mutations within the cytoplasmic domain of the CD4 molecule that severely impair endocytosis have no effect on HIV infectivity (Bedinger et al. 1988; Maddon et al. 1988). Moreover, the cytoplasmic and membrane-spanning domains of CD4 are dispensable for HIV receptor function. Molecules with a truncated cytoplasmic domain, or those in which these two regions have been substituted by foreign anchor/cytoplasmic domains or even glycophospholipid anchors, can act as receptors for HIV entry into human cells (Bedinger et al. 1988; Diamond et al. 1990; Kost et al. 1991; Golding et al. 1993; Marshall et al. 1994). Thus, as with many other viral systems, the machinery for virion entry is provided by the virus, not the host.
Molecularly engineered forms of the CD4 molecule that lack the hydrophobic anchor domain and are secreted from the cell, known as soluble CD4 (sCD4), bind efficiently to gp120 or intact virus, even when the truncation leads to secretion of just the first two immunoglobulinlike domains (Berger et al. 1988; Richardson et al. 1988). Fusion of these domains to an amino-terminally truncated CD8 molecule provides a functional, if somewhat less efficient, receptor for HIV that can mediate both cell-cell fusion and viral entry (Poulin et al. 1991; Golding et al. 1993).
The Importance of Second (or Fusion) Receptors for HIV Infection
The primate lentiviruses appear to differ from all other retroviruses in the specific requirement for a second cell surface molecule for infection. In vivo, the major cell types infected by HIV are those expressing CD4. These include the T-helper lymphocytes, cells of the monocyte-macrophage lineage, and dendritic, antigen-presenting cells (for review, see Weiss 1993; see Chapter 11). Most viral isolates obtained from infected individuals exhibit one of two distinct patterns of infectivity (tropism). All isolates of HIV-1 replicate well in vitro in activated CD4+ T cells present in peripheral blood mononuclear cell (PBMC) cultures. Some, but not all, can also replicate in mature macrophage cultures. These macrophage (or M)-tropic isolates are generally unable to replicate in established CD4+ T-cell lines. Other isolates, particularly those obtained at late stages of infection, or adapted to grow in permanent cell lines, replicate to high titers both in PBMC and in T-cell lines but are usually unable to grow in macrophages. These strains can also show differential growth properties on different T-cell lines. T-cell line (or T)-tropic variants of HIV also exhibit a greater propensity to induce cell fusion in culture (see below) and are often referred to as syncytium-forming (SI) isolates. Rarely, isolates that display a “dual” tropic phenotype, capable of replicating in both T-cell lines and macrophages, can be found (Collman et al. 1992; Shibata et al. 1995). Molecularly cloned proviruses encode viruses that retain these varying properties, showing them to be heritable traits defined by the virus.
Although CD4 is necessary and sufficient for efficient HIV attachment to cells, its presence alone is clearly not sufficient for viral entry, since not all human cells engineered to express CD4 are susceptible to infection (Chesebro et al. 1990b; Clapham et al. 1991). Furthermore, transfection of a variety of mammalian cells with the cDNA for human CD4 allows gp120 binding, but neither viral entry nor fusion with env-expressing cells occurs (Maddon et al. 1986; Clapham et al. 1991). Although a variety of nonhuman cells expressing Env protein can fuse with CD4-expressing cells, this can occur only if the latter cells are of human origin (Ashorn et al. 1990; Aoki et al. 1991). Thus, the block to fusion is determined specifically by the cell expressing the CD4 receptor. Moreover, whereas HIV-1 virions and VSV pseudotypes bearing the HIV-1 glycoprotein complex are unable to infect a variety of CD4-expressing mammalian cells, bound virus can be induced to initiate infection following treatment with polyethylene glycol (PEG) (Clapham et al. 1991). Since HIV-1 can productively infect a variety of nonhuman cells when it is pseudotyped with the envelope proteins of amphotropic or xenotropic MLV (Canivet et al. 1990; Chesebro et al. 1990a; Page et al. 1990; Spector et al. 1990), it seems unlikely that postfusion (i.e., uncoating) events in the entry process are defective in nonpermissive cells. Thus, the block to entry in these cells appears to be after binding but prior to fusion. One possibility for such a block could be the inability of CD4 alone to induce the necessary conformational changes in the glycoprotein complex that are required for this process to occur. The demonstration that both stable CD4+ mouse-human hybrids containing only a subset of human chromosomes (Weiner et al. 1991; Ramarli et al. 1993) and CD4+ human-mouse heterokaryons (Dragic et al. 1992; Broder et al. 1993) are susceptible to viral infection and fusion by HIV gp120 points to the absence of a critical factor for HIV-1 entry, rather than the presence of an inhibitory molecule in the nonpermissive cells.
In response to the above observations, a number of candidates were proposed for the accessory molecules that in addition to CD4 define permissiveness for HIV. These included the cell surface adhesion molecule LFA-1 (CD18) (Hildreth and Orentas 1989; Valentin et al. 1990), tryptase TL2, a membrane-bound serine esterase (Hattori et al. 1989; Clements et al. 1991), and the cell surface diaminopeptidase CD26 (Callebaut et al. 1993). However, results of additional experimental approaches did not support these as cofactors for entry of HIV in CD4+ cells (Pantaleo et al. 1991; Alizon and Dragic 1994; Broder et al. 1994; Camerini et al. 1994; Lazaro et al. 1994; Patience et al. 1994).
Recent studies have led to the definitive identification of at least two second receptor molecules and, in the process, have provided new insight into a number of previously rather puzzling aspects of HIV infection. Feng et al. (1996) identified a clone from a human cDNA library capable of making mouse cells expressing human CD4 permissive for fusion with cells expressing HIV Env protein. The gene represented by this clone, called “fusin” (also LESTR) for its ability to promote HIV-induced cell fusion is now called CXCR4, for its role as a receptor for the CXC class of chemokines. It encodes a transmembrane glycoprotein of about 46 kD, with seven membrane-spanning segments predicted from the amino acid sequence (Fig. 5). CXCR4 belongs to a family of receptors that signal through interaction with G proteins and is most closely related to the receptor for the α-chemokine interleukin-8 (IL-8); its ligand has recently been identified as SDF-1, a stromal-derived factor that serves as a chemoattractant for lymphocytes (Bluel et al. 1996; Oberlin et al. 1996). When coexpressed with human CD4, it confers on mouse cells not only the ability to fuse with env - expressing cells, but also the ability to be infected by T-tropic (see below) strains of HIV. Its role as an essential cofactor in HIV infection is supported by the ability of antibodies against its amino terminus to block infection of T-cell lines, as well as the fact that expression of CXCR4 in human cell lines coincides with their infectability.
The specificity of CXCR4 for T-cell-tropic HIV as well as the recently discovered role of β-chemokines in suppressing HIV infection (Cocchi et al. 1995; see below) suggested that a related protein might be involved in infection of cells by M-tropic virus as well. This hypothesis was confirmed with the observation that a related receptor protein, called CCR5, confers on CD4-expressing cells the ability to be infected by M-tropic strains of HIV (Table 5) (Alkhatib et al. 1996; Deng et al. 1996; Doranz et al. 1996; Dragic et al. 1996). Unlike fusin, the ligands for CCR5 are known. They include a set of β-chemokines, including RANTES (regulated upon activation, normal T-cell expressed and secreted), MIP-1α (macrophage inflammatory protein-1α), and MIP-1β. Chemokines are a large family of small proteins secreted by many cell types that serve as chemoattractants to draw cells to sites of inflammation. As with fusin, the pattern of expression of CCR5 on CD4-expressing cells reflects their susceptibility to infection by M-tropic HIV strains, and joint expression of the chemokine receptor and human CD4 seems to render any cell line—even quail cells—susceptible to infection by most but not all strains. The use by some of the dual-tropic isolates of both receptors, as well as the related CCR3 and CCR2b (Choe and Sodroski 1995; Doranz et al. 1996), suggests that there may be a larger family of second receptors controlling subtle differences in tropism. The existence of other molecules acting as second receptors is also implied by the different host ranges of HIV-2 and SIV (Table 6).
Table 5
Role of Fusin and CC-CKR5 in HIV Infection.
Table 6
Differential Infection of CD4+ Cells Lines by HIV-1, HIV-2, and SIV.
Consistent with a role for the CCR5 chemokine receptor as a coreceptor for HIV, pretreatment of cells expressing the receptor with its chemokine ligands (RANTES, MIP-1α, and MIP-1β) blocks infection with M-tropic, but not T-cell-line-tropic HIV-1 (Alkhatib et al. 1996; Deng et al. 1996; Dragic et al. 1996). Remarkably, these proteins had been identified previously as components of the inhibitory factors released by activated CD8+ T cells which are responsible for suppressing HIV replication in mixed cultures of CD4 and CD8 cells (Cocchi et al. 1995) (see Chapter 11). Their importance in regulating HIV infection in vivo is implied by the correlation between high levels of chemokine expression and resistance of certain individuals to infection by HIV despite repeated exposure (Dragic et al. 1996; Paxton et al. 1996), as well as the presence in a large fraction of HIV-exposed but uninfected individuals of a mutant form of CCR5 with a 32-base deletion causing a frameshift and complete failure to synthesize active receptor. Individuals homozygous for this mutation constitute about 10% of Europeans and much lower fractions of Asians or Africans. They appear to be completely resistant to infection by HIV-1 by any route. Heterozygotes are susceptible to infection but may have a reduced rate of progression to disease (Dean et al. 1996; Liu et al. 1996; Paxton et al. 1996).
Alternate Receptors for HIV
Since the CD4 molecule was at one time thought to be found only on T cells, infection of macrophages, monocytes, and related cells was believed to be independent of CD4. However, inhibition of infection by anti-CD4 monoclonal antibodies (Collman et al. 1990) demonstrated that the low levels of CD4 present on these cells were necessary for HIV infection. In contrast, a variety of human cell types, primarily of nervous system, endothelial, and fetal origins, do not express CD4 but are sensitive to HIV infection (for review, see Clapham et al. 1993; Weiss 1993). In general, however, the efficiency of infection is very low, and a large dose of input virus is required to yield a small number of infected cells. Nevertheless, infection of these cells cannot be inhibited by soluble CD4 or by monoclonal antibodies that compete for the gp120-binding site, which points to the presence of alternative, albeit less efficient, receptors for HIV entry. One possible candidate for such a receptors is the glycolipid galactosyl ceramide (galactocerebroside, or GalC). Harouse et al. (1991) showed that antibodies to GalC could block HIV-1 infection of two CD4-negative cell lines derived from the nervous system and that recombinant gp120 could bind to the glycolipid with an affinity approaching that of CD4. The same molecule appears to mediate entry of HIV-1 into the colon cancer cell line HT-29; the efficiency by which subclones of these cells could be infected correlates with the level of GalC on their surfaces (Yahi et al. 1992; Fantini et al. 1993). The binding site for GalC is clearly distinct from that of CD4 since sCD4 cannot block binding of the glycolipid to gp120; however, attempts to map the site have led to different results (Bhat et al. 1993; Cook et al. 1994).
A discrete alternate receptor appears to mediate entry of certain HIV-2 strains into some cell lines that lack CD4. Clapham et al. (1992) showed that a variant of HIV-2 ROD (ROD/B) infects RD/TE671 rhabdomyosarcoma cells and the B-cell lines, Raji and Daudi, with an efficiency approaching that of the highly permissive CD4+ cell line C8166. Most other HIV-2 strains tested were either much less efficient or totally unable to infect the CD4-negative cell lines, but they could be induced to infect and mediate fusion of these cells following incubation with sCD4. Thus, it appears that the RD, Raji, and Daudi cells express a cell surface molecule recently identified as CXCR4 (Endres et al. 1996) that can act as a receptor for HIV-2 ROD/B and that most strains of HIV-2 require activation by sCD4 in order to undergo a productive (fusogenic) interaction with the coreceptor. Weiss (1993) has suggested that the HIV-2 ROD/B variant might adopt this “fusogenic conformation” in the absence of sCD4.
Activation of HIV-2 for entry into CD4-negative cells seems to be similar in principle to sCD4 enhancement of SIVAGM entry into human CD4+ T-cell lines reported by Allan et al. (1990) and Werner et al. (1990). It is therefore possible that in both cases, conformational changes induced by sCD4 allow gp120 or gp41 interactions with a common alternate receptor on the cell surface. Whether interactions with such a molecule could be a prerequisite step in the normal process of CD4+ cell entry by HIV-1, HIV-2, or SIV remains to be determined.
Incompletely Characterized Receptors for Other Retroviruses
Mammalian Type C
Feline Type-C Retroviruses
Based on neutralization and interference properties, feline leukemia virus (FeLV) isolates can be classified into three subgroups (A, B, and C) (Jarrett et al. 1973; Sarma et al. 1975). Host range parallels the subgroup classification: Subgroup A viruses have a restricted, ecotropic host range, whereas subgroup B and C viruses have a more extended host range. Subgroup B viruses are frequently found in association with those of subgroup A and probably arise as the result of recombination between an infecting subgroup A virus and endogenous proviruses in the cat genome (Elder and Mullins 1983; Overbaugh et al. 1988; Sheets et al. 1992). Even though their envelope gene sequences are highly related to those of subgroup A, the subgroup C viruses can also infect cells other than of cat origin. It seems likely that they arise by mutation from the subgroup A viruses rather than by recombination (Neil et al. 1991). The FeLV-B receptor was discussed above; the others remain to be identified.
Primate Type-C Viruses
Baboon endogenous virus (BaEV) was initially isolated by cocultivation of a normal baboon placenta with various cell lines (Benveniste et al. 1974; see Chapter 8). As pointed out above, evidence from interference and host-range studies has indicated that BaEV and members of the RD114/CCC group of feline endogenous viruses utilize the same receptor for cell entry; the two groups are also related at nucleic acid and serological levels. As with the murine endogenous, xenotropic viruses, BaEV is unable to infect baboon cells, but it can infect human, rhesus monkey, dog, and bat cells. Although none of the isolates replicate in cat, mink, rat, mouse, or avian cells, at least in some of these species, the block may be at a postentry stage.
BaEV/RD114 viruses form part of an unusual, diverse grouping of retroviruses from different genera which utilize the same receptor. The glycoproteins of the BaEV/RD114 group are related to both those of the primate type-D group and those of the avian reticuloendotheliosis virus (REV) group, and it seems likely that the env genes of these viruses were derived from a common progenitor. Despite the sequence divergence in their gp70 (SU) proteins, they clearly share functional, receptor-binding determinants, since complete viral interference is observed between the different groups. Cells infected by a D-type virus are resistant to infection by BaEV or RD114 and by VSV or MSV pseudotypes of these viruses (Sacks et al. 1978; Chatterjee and Hunter 1980; Sommerfelt and Weiss 1990). Similarly, REV infection is prevented by preinfection of cells with D-type simian retroviruses (Kewalramani et al. 1992). Given the large evolutionary distance between these genera and the extensive divergence in their SU glycoproteins, the conservation of a common receptor-binding site suggests that switching receptors is a rare and stochastic event. The molecular nature of the receptor for this broad grouping of viruses remains to be established.
A second class of endogenous primate type-C retroviruses is represented by the MAC-1/MMC-1 isolates from macaques and the related isolate from the colobus monkey, CPC-1. Like BaEV, these viruses are xenotropic, unable to infect the species from which they were isolated, but able to infect human cells in culture. In contrast to BaEV, they can also infect cat cells but do not appear to be able to infect bat, dog, mink, or mouse cells, and so the receptor for this group of viruses is likely to be distinct from that of the BaEV/RD114 group (Teich 1984).
Reticuloendotheliosis Viruses
In addition to the well-characterized ASLV group, retroviruses isolated from birds include the REVs, which are classified with the mammalian C-type viruses. Both nucleic acid and protein studies have shown that REV is strikingly similar to retroviruses of primates (Sonigo et al. 1986). As described above, REV infection is prevented by preinfection of cells with D-type simian retroviruses; i.e., these viruses exhibit receptor interference (Kewalramani et al. 1992). Thus, it appears likely that the glycoproteins of REV and the D-type retroviruses, as well as some primate type-C viruses, share a common receptor.
D-type Retroviruses
In 1970, a virus with many characteristics of mouse mammary tumor virus was isolated from a mammary tumor of a rhesus monkey (Chopra and Mason 1970). This virus (Mason-Pfizer monkey virus, MPMV) is a horizontally transmitted exogenous virus of the rhesus monkey and represents the prototype virus of the D-type retroviruses, a group that includes endogenous viruses of the langur and squirrel monkeys and an increasing number of exogenous immunosuppressive isolates from macaque species (Daniel et al. 1984; Marx et al. 1984, 1985; Stromberg et al. 1984). This group of viruses has a fairly narrow host range, primarily infecting macaque, human, and dog cells (for review, see Fine and Schochetman 1978). However, there appears to be little tissue specificity for infection, since these viruses can infect fibroblasts and lymphoid cells of both T-cell and B-cell lineage. Sommerfelt and Weiss (1990) have shown that all of the primate D-type isolates utilize a common receptor—infection of cells by any member can establish interference to entry by each of the remaining members. As described above, cells infected by a D-type virus also exhibit interference to infection by BaEV and the related RD114 (Chatterjee and Hunter 1980; Sommerfelt and Weiss 1990), consistent with a common receptor. The basis of this phenomenon may lie in a common origin for the env gene of these divergent species. Since the gag-pol region of MPMV appears to have been derived from a common progenitor with the murine B-type viruses (and IAPs), it seems likely that the primate type-D family arose by recombination of such sequences with a primate type-C env gene (Sonigo et al. 1986).
Mouse Mammary Tumor Virus
The mouse mammary tumor viruses (MMTVs) are B-type retroviruses. Like the primate D-type retroviruses, this family assembles immature capsids (referred to historically as intracytoplasmic A-type particles) intracellularly. As their name implies, MMTVs are most frequently associated with the induction of breast cancer in mice. MMTV is generally transmitted horizontally, as an infectious virus in the milk of infected females, or vertically, as an endogenous provirus. It is difficult to establish de novo infection of cells in culture with MMTV, even though experiments with VSV pseudotyped with the MMTV envelope glycoprotein (Fig. 1) have shown that many cell types express the receptor (Zavada et al. 1977; Chan et al. 1982). Both MMTV and VSV(MMTV) pseudotypes can infect cells of nonmurine species (Lasfargues et al. 1976; Zavada et al. 1977). Moreover, studies by Altrock et al. (1981), using pseudotypes of MSV, suggested that distinct ecotropic and xenotropic variants of MMTV exist.
The genetic determinant for susceptibility to ecotropic MMTV has been mapped to chromosome 16 by using the VSV(MMTV) pseudotype virus to infect a panel of mouse–Chinese hamster cell hybrids (Hilkens et al. 1983); however, a molecular characterization of the receptor remains to be performed.
Human/Simian T-cell Leukemia Virus (HTLV-1/2), Bovine Leukemia Virus
HTLV-1 can bind to and infect a wide variety of different cell types in vitro (Clapham et al. 1983; Nagy et al. 1983; Krichbaum-Stenger et al. 1987), although only T and B cells are susceptible to virus-induced transformation (Chen et al. 1984). The in vivo T-cell tropism of the virus is therefore not at the level of receptor expression; rather, postpenetration events appear to determine HTLV host range. Although in vitro infection of nonlymphoid cells has been achieved in human osteosarcoma cells (HOS) and human tumor cells (HT1080), it in not clear whether nonlymphoid cells are infected in vivo.
In addition to binding assays, syncytial and pseudotype assays have been used to determine functional receptor gene expression and HTLV host range. The results of such approaches indicate that HTLV-1 and HTLV-2 share the same cell surface receptor (Weiss et al. 1985) and the receptor gene for both HTLV-1 and HTLV-2 has been assigned to human chromosome 17 (Sommerfelt et al. 1988). The molecular identity of its product is currently unknown.
BLV, a naturally occurring exogenous B-cell lymphotropic retrovirus, is the etiologic agent of enzootic bovine leukosis, a disease characterized by an initial persistent lymphocytosis and then by the occurrence of clonal lymphoid tumors of B-cell origin (Ghysdael et al. 1985). BLV infects a variety of cells in vitro and can propagate in various animal species. This bovine retrovirus and HTLV-1 and HTLV-2 are related retroviruses which share a common genome organization, presence of the regulatory proteins Tax and Rex, and nucleotide sequence similarity. However, because BLV and the HTLVs do not infect the same target cells, they appear to utilize different cell receptors. By screening a cDNA expression library derived from a permissive bovine cell line (MDBK) for a clone that could bind soluble gp51(SU), Ban et al. (1993) identified a candidate receptor gene for BLV. NIH-3T3 cells transfected with the gene and expressing detectable amounts mRNA showed increased susceptibility to BLV infection (Ban et al. 1993). On the basis of its sequence, the protein encoded by this gene appears to be a class-1 membrane glycoprotein of about 70 kD with no remarkable structural features or relationship to known proteins. Its function is also unknown.
Spumaviruses
Retroviruses within this family are commonly referred to as foamy viruses, because of the characteristic vacuolated “foamy,” degeneration that they induce in cells in culture. They are present in a number of mammalian species where they establish persistent infections without evident pathogenesis in their natural hosts. These viruses are amphotropic, infecting cells of epithelial, fibroblastic, and lymphoid origins in culture from a wide range of species including human, monkey, pig, rabbit, mouse, rat, mink, cat, dog, and chicken (Hooks and Gibbs 1975). Even cell lines from more exotic species (e.g., dolphin, monkjack, and iguana), in addition to the more frequently used animal and rodent cell lines, are readily infectible by the virus, suggesting that its receptor is widely distributed on all cell types from many species (McClure and Erlwein 1995).
Nonprimate Lentiviruses
The nonprimate lentiviral group includes the prototype “slow virus” visna/maedi virus (VMV), as well as the related caprine arthritis-encephalitis virus (CAEV), equine infectious anemia virus (EIAV), and the more recently described feline and bovine immunodeficiency viruses (FIV and BIV).
VMV replicates in the cells of many vertebrate species, but viral production is most efficient in ovine cells, particularly in cell cultures of sheep choroid plexus. CAEV replication, in contrast, is restricted to goat cells and occurs most efficiently in goat cultures of synovial membranes (mostly macrophages). Results consistent with neutralization studies suggest that the env gene products of these two viruses are distinct (Teich 1984). EIAV shows a similarly restricted host range for replication, targeting primarily macrophages in vivo and replicating most efficiently in these cells in vitro. In studies of CAEV and EIAV, however, it is not clear whether the block to viral replication is because receptors are lacking or because a postpenetration function is defective. More detailed studies aimed at defining the nature of the receptors and their tissue and species distribution for these viruses are clearly necessary.
FIV is a widespread pathogen of domestic cats, and infection with the virus causes an immunological dysfunction that resembles AIDS in humans. Although the cell tropism of FIV is similar to that of HIV (T cells, monocytes/macrophages), a poor correlation exists between feline CD4 expression and susceptibility to FIV infection in vitro (Willett et al. 1991), implying the use of a protein other than CD4 as the receptor. Recent work, using monoclonal antibodies to feline cell surface molecules, has identified the feline CD9 homolog as a potential receptor for this virus (Hosie et al. 1993; Willett et al. 1994). Unlike CD4, CD9 is expressed on a wide variety of cells and seems to be important in nervous system development. If, indeed, CD9 is its principal receptor, the specificity of FIV infection in vivo must reside in factors other than receptor distribution.
- Receptors - RetrovirusesReceptors - Retroviruses
Your browsing activity is empty.
Activity recording is turned off.
See more...