Except where otherwise noted, this work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC-BY-NC-ND 4.0). To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
Preface
Molecules on the surface of S. pyogenes range from a complex coiled coil modular structure, like the M protein, to conventional globular proteins and polysaccharides. All of the molecules on the bacterial surface have specifically evolved to enable survival in its human environment like the throat, blood, or skin. In particular, the M protein molecule has been finely tuned to allow the streptococcus to persist in infected tissues while skillfully avoiding human immune cells. As the first molecule on S. pyogenes to be completely sequenced, it is considered the archetypical surface molecule for these organisms and other Gram-positive bacteria; and as a result, in this chapter, it will be used as a comparator for the other surface proteins on streptococci.
Introduction
Streptococcus pyogenes (or group A streptococcus) is responsible for a number of suppurative human infections, of which acute pharyngitis and impetigo are the most common. As a consequence of ineffective antibiotic therapy or no therapy, as many as 3 to 5% of individuals who suffer a group A streptococcal pharyngeal infection may develop acute rheumatic fever (Wannamaker, 1973; Breese, 1978), a disease that often results in cardiac damage, particularly to the mitral valve. Rheumatic fever is not currently a major problem in the developed world; however, it is the leading cause of heart disease in school-age children in developing nations (Agarwal, 1981). Recent estimates indicate that there are at least 517,000 deaths worldwide each year due to group A streptococcal diseases (such as acute rheumatic fever, rheumatic heart disease, acute post-streptococcal glomerulonephritis, and invasive infections) (Carapetis, Steer, Mulholland, & Weber, 2005). Rheumatic heart disease, with a prevalence of at least 15.6 million cases and 233,000 deaths each year, has the greatest disease burden (Rammelkamp & Weaver, 1953).
The ability of S. pyogenes to persist in infected tissues can be primarily attributed to the cell surface M protein, a molecule that gives the streptococcus the ability to resist phagocytosis by polymorphonuclear leukocytes in the absence of type-specific antibodies. The importance of the M molecule is seen in an M-knockout mutant, which has all the other surface and secretory molecules of the wild-type organism, yet will not survive in human blood that contains phagocytes (Perez-Casal, Caparon, & Scott, 1992). Resistance to a group A streptococcal infection is directly related to the presence of type-specific antibodies to the M molecule (Lancefield, 1959; Lancefield, 1962). Since there are >200 different serotypes of M protein (i.e., M6, M12, M18, M24, and so on), an individual may become infected by more than one group A streptococcal type during a lifetime (Lancefield, 1962). Rebecca Lancefield identified the streptococcal M protein nearly 90 years ago (Lancefield, 1928). A review by Lancefield in 1962 (Lancefield, 1962) clearly describes the studies carried out over a 35-year period that defined this molecule as a major virulence factor for the Streptococcus pyogenes bacterium.
The streptococcal M protein is now probably one of the best-defined molecules among the Gram-positive bacterial virulence determinants. Its structure, function, immunochemistry, and method of antigenic variation are unique among known virulence molecules, and may serve as a model for certain microbial systems (Fischetti, 1989).
Streptococcal Cell Wall
Like the cell wall of other Gram-positive bacteria, the streptococcal cell wall contains a thick peptidoglycan, which is intercalated and covered with protein, teichoic acid, and lipoteichoic acid. The group-specific antigenic determinant for the group A streptococcus, which differentiates it from other streptococci, is the β-linked N-acetylglucosamine, which caps the polyrhamnose chains that extend from the N-acetylmuramic acids of the peptidoglycan (Coligan, Schnute, & Kindt, 1975). In contrast, group C streptococci (which cause human and animal infections) has an α-linked N-acetylgalactosamine as its group-specific determinant (Coligan, Schnute, & Kindt, 1975; Coligan, Kindt, & Krause, 1978). Surface proteins on streptococci contain an N-terminal signal peptide for Sec-dependent secretion and may be separated into three main categories: those that anchor at their C-terminal end (through an LPXTG motif); those that bind by way of charge or hydrophobic interactions; and those that bind via their N-terminal region (or lipoproteins, which are characterized by a cysteine-containing 'lipobox' within their signal peptide sequence) (Sutcliffe & Harrington, 2002) (Figure 1).
M Protein
M protein is the major virulence determinant for S. pyogenes, since M- mutants are unable to survive in phagocyte-containing human blood (Maxted, 1956), thus all human isolates have surface M-protein, which appears like the fuzz on a tennis ball when viewed by transmission electron microscopy (Figure 2). It was subsequently found that protection against infection by these organisms was dependent on the presence of antibodies directed to the N-terminal region of the M protein (Cunningham & Beachey, 1974; Beachey, Campbell, & Ofek, 1974), the type-specific segment of the molecule (Beachey, Seyer, Dale, Simpson, & Kang, 1981; Dale, Seyer, & Beachey, 1983). It is known that the region of the M molecule responsible for this type specificity is limited to 50 or so amino acids on the N-terminus. However, the mechanism by which the sequence in this region changes to produce a new serotype is unknown.
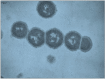
Figure 2.
Thin section electron micrograph of a chain of streptococci showing the surface M protein. Magnification 50,000x.
M protein structure
M protein may be considered the archetypical molecule for those surface proteins that anchor via their C-terminal region on the Gram-positive bacterial cell wall. A feature of the M molecule, as with many surface proteins on bacteria, is its multi-domain structure. As Figure 3 shows, the M6 protein is composed of four sequence repeat domains, with each differing in size and sequence (Hollingshead, Fischetti, & Scott, 1986). The A-repeats are each composed of 14 amino acids, where the central blocks are identical and the end blocks slightly diverge from the central consensus repeats. The B-repeats, composed of 25 amino acids each, are arranged similar to the A-repeats. The C-repeats, composed of 2.5 blocks of 42 amino acids each, are not as identical to each other as the A and B repeats. There are also 4 short D-repeats, which show some homology among each other. These repeat segments make up the central helical rod region of the M6 and other M molecules, because of the high helical potential ascribed to the amino acids found within this region, as determined by conformational analysis (Fischetti, et al., 1988; Phillips, Flicker, Cohen, Manjula, & Fischetti, 1981). Recombination within the repeats has been shown to cause size variation among and within M proteins of the same and different serotypes, even in strains isolated from the same outbreak (Fischetti, Jones, & Scott, 1985; Fischetti, Jarymowycz, Jones, & Scott, 1986; Hollingshead, Fischetti, & Scott, 1987). It has been suggested that this could be a strategy by which the organism could escape immune recognition.
Examination of the sequence within the helical rod region of the M6 molecule revealed a repeating 7-residue periodicity of non-polar amino acids, a characteristic of α-helical coiled-coil proteins like mammalian tropomyosin and myosin. Generally, α-helical coiled-coil molecules are constructed from a reiterating 7-residue pattern of amino acids (a-b-c-d-e-f-g)n where residues in positions 'a' and 'd' are hydrophobic and form the 'core' residues in the coiled-coil, while the intervening residues are primarily helix-promoting. Irregularities in the heptad pattern, which are found especially in the B repeat region and other coiled-coil molecules, probably account for the flexibility of the M molecules that has been observed in electron micrographs (Phillips, Flicker, Cohen, Manjula, & Fischetti, 1981). Based on these discontinuities in the heptad pattern, the central rod region is divided into three sub-regions, which correlate to the A-, B-, and C-repeat blocks (Fischetti, 1989).
Whereas M6 and other M molecules conform to this general arrangement of amino acids, other M proteins diverge somewhat from this configuration. A recent analysis of 1086 streptococcal isolates from 31 countries, representing 175 M protein types, revealed differences in the central rod region of the molecule showing fewer repeats, in some cases, and a shorter rod as a result (McMillan, et al., 2013). It was found that the A-repeats, in particular, were missing from the majority of the M proteins, which belonged to pattern D and E M proteins (see the chapter in this book on the molecular basis of stereotyping for more information).
Despite several attempts to crystallize the whole M molecule, only an N-terminal fragment that contains both the A and B repeats of the M1 protein was successfully accomplished (McNamara, et al., 2008). The structure confirmed the presence of significant irregularities in the coiled-coil structure also found in other mammalian coiled-coil proteins, like tropomyosin and myosin. The authors found that if these irregularities were corrected in the B-repeat region (a region responsible for fibrinogen binding), a loss in fibrinogen binding occurred, which suggests that these irregularities are biologically important. The high structural similarity between the M and host α-helical coiled-coil proteins has led researchers to hypothesize the elicitation of anti-M cross-reactive antibodies and T cell receptors directed against these human proteins as a possible cause for S. pyogenes-associated autoimmune sequelae (Cunningham, 2000).
Swanson et al. (Swanson, Hsu, & Gotschlich, 1969) published the first electron micrographs (EM) of the M protein on the surface of S. pyogenes. This was the first evidence to show that the M molecule was an elongated structure. Although at the time, other proteins were reported to also be on the streptococcal surface, they were not apparent in these EM preparations. Thus, direct visualization of surface molecules is limited to those with certain physicochemical characteristics. In the case of the M protein, it is the α-helical coiled-coil structure that allowed for this visualization (Figure 2 & 4).
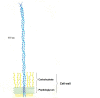
Figure 4.
The appearance of the M molecule on the surface of the streptococcal cell wall. See caption of Figure 3 for more details.
In experiments designed to answer questions about the synthesis and placement of M protein on the cell wall, Swanson et al. (Swanson, Hsu, & Gotschlich, 1969) found that after trypsinization to remove existing M protein on living streptococci, newly synthesized M protein was first seen by EM on the cell in the location of the newly forming septum. In similar experiments designed after the classical experiments of Cole and Hahn (Cole & Hahn, 1962), trypsinized streptococci placed in fresh media for ten minutes revealed M protein first at the newly forming septum (Raz, Talay, & Fischetti, 2012); whereas Sfb (a fibronectin binding protein on streptococci) first appears at the poles and increases gradually in concentration, as compared to its rapid appearance of the M molecule. On organisms examined after 40 minutes or more of incubation, M protein was not observed in the position of the “old wall,” which suggests that the M molecule is produced only where new cell wall is synthesized, and confirms the observations of Swanson et al. (Swanson, Hsu, & Gotschlich, 1969). These results demonstrate a close relationship between the regulation of cell division and protein anchoring (for more information, refer to the chapter on the spatial regulation of protein sorting in this book).
C-terminal anchor region
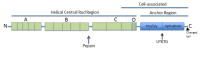
Hundreds of proteins from Gram-positive bacteria have now been reported that anchor to the cell wall through their C-terminal region (Table 1 shows a representative list of streptococcal surface proteins). From sequence alignments of a variety of surface proteins on Gram-positive bacteria, it became obvious that there was a common theme within the C-terminal region of these molecules, without exception: all had a similar arrangement of amino acids (Figure 5). Up to seven charged amino acids are found at the C-terminus, which are composed of a mixture of both negatively and positively charged residues. Immediately N-terminal to this short-charged region is a segment of 15-22 predominately hydrophobic amino acids that are sufficient to span the cytoplasmic membrane of the bacterium. In all these proteins, the sequences found in the hydrophobic and charged regions are not necessarily identical, but the chemical characteristics of the amino acids used to compose them are conserved. About eight amino acids N-terminal from the hydrophobic domain is a heptapeptide with the consensus sequence LPXTG, which is extraordinarily conserved among all the C-terminal-anchored proteins examined (Fischetti, Pancholi, & Schneewind, 1990). While several amino acid substitutions are seen in position 3 of the heptapeptide (predominantly A, Q, E, T, N, D, K, and L), positions 1, 2, 4, and 5 are nearly 100% conserved (see Table 1). This conservation is also maintained at the DNA level. The preservation of this hexapeptide and the high homology within the hydrophobic and charged regions suggest that the mechanism of anchoring these molecules within the bacterial cell is also highly conserved.
Table 1:
C-terminal LPXTG-Linked Surface Proteins on Streptococci
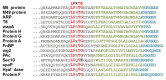
Figure 5.
Sequence alignment of the C-terminal end of M protein and other streptococcal surface proteins, depicting the charged tail (blue), hydrophobic domain (green), and LPXTG motif (red). Sequences are aligned at the LPXTG motif.
The secretion and anchoring of surface proteins are highly regulated in S. pyogenes. For example, as previously mentioned, anchoring M-protein to the cell wall occurs exclusively at the septum (Cole & Hahn, 1962; Swanson, Hsu, & Gotschlich, 1969), along with the biosynthesis of the cell wall (Raz & Fischetti, 2008). The resulting coupling of M-protein anchoring and cell wall synthesis leads to the coating of the entire cell surface with M protein. In contrast to M-protein, SfbI (also known as protein F, or PrtF), a major fibronectin-binding protein in certain streptococcal strains (Talay, Valentin-Weigand, Jerlström, Timmis, & Chhatwal, 1992a; Hanski & Caparon, 1992), is anchored at the old poles (Ozeri, et al., 2001; Raz & Fischetti, 2008). Secretion of M-protein and SfbI at their respective cellular locations is directed by information found in their signal sequences (Carlsson, et al., 2006). Proteins that contain a YSIRK/GS motif in their signal sequence are directed for secretion at the septum, while proteins that do not contain this motif are secreted at the poles, or in a hemispherical distribution. The N-terminal signal sequence directs these molecules for translocation through the secretion channel, and translocation is halted when the C-terminal LPXTG sorting signal reaches the channel. At this point, the LPXTG motif is exposed on the extracellular side of the membrane, the hydrophobic stretch spans the membrane, and the C-terminus positively-charged residues are within the cytoplasm (Figure 6) (Schneewind, Mihaylova-Petkov, & Model, 1993; Schneewind, Pancholi, & Fischetti, 1992). At this time, the membrane-associated transpeptidase sortase then cleaves the LPXTG motif between the threonine and glycine residues, and connects the freed threonine to lipid II (Marraffini & Schneewind, 2006; Mazmanian, Ton-That, & Schneewind, 2001a; Perry, Ton-That, Mazmanian, & Schneewind, 2002). The lipid II–protein complex is then processed by penicillin binding proteins and finally attached to the cell wall. The C-terminal portion of the cleaved sorting signal, which contains the hydrophobic region and positively charged residues, is released back into the cytoplasm (Navarre & Schneewind, 1994).
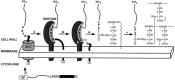
Figure 6.
Translocation of proteins with an LPXTG motif to the surface of the cell wall for anchoring (Mazmanian, Ton-That, & Schneewind, 2001b).
Wall-Associated Region
Immediately N-terminal from the LPXTG motif is the wall-associated region, which spans from about 50 to as many as 125 amino acid residues, and is found in nearly all C-terminal anchored surface proteins analyzed. Although this region does not exhibit a high degree of sequence identity among the known proteins, it is characterized by a high percentage of proline/glycine and threonine/serine residues. For some proteins (like the M protein), the concentration of proline/glycine is significantly higher than threonine/serine, while in others this relationship is either reversed or nearly equal. Because of its proximity to the hydrophobic domain, which is imbedded in the cell membrane, this region would be positioned within the peptidoglycan layer of the cell wall (Wannamaker, 1973), (Figure 4). The reason for the presence of these particular amino acids at this location is unknown. One hypothesis suggests that the prolines and glycines, with their ability to initiate bends and turns within the secondary structure of proteins, allow the peptidoglycan to more easily become cross-linked around these folds, which further stabilizes the molecule within the cell wall (Pancholi & Fischetti, 1988). The function of the threonines and serines within this region is not immediately apparent. While these amino acids are commonly used as O-linked glycosylation sites in eukaryotic proteins, such substitutions have not yet been established in this region of surface molecules.
Surface Exposed Region
As the C-terminal region is characteristically conserved among the various streptococcal surface proteins, the regions exposed on the cell surface are characteristically unique. Despite their differences, these molecules appear to fall into three groups: i) those with several domains; ii) those with repeat domains located close to and within the cell wall region; and iii) those without any repeat domains (Figure 7). Streptococcal M protein and protein G may be considered representative molecules of those containing several sequence repeat domains. However, the most commonly found structures are those with repeats located close to the cell wall and an extended non-repetitive region N-terminal to this segment.
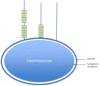
Figure 7.
Conformational characteristics of the proteins found on the surface of streptococci. Molecules with multiple repeats (like M protein), molecules with few repeats (usually located close to the wall), and molecules with no repeats are found.
Conformationally analyzing 12 representative surface molecules through the algorithm of Garnier et al. (Garnier, Osguthorpe, & Robson, 1978) revealed that those proteins that contain repeat sequences were predominantly helical within the region containing repeat segments (Figure 7). On the other hand, regions and molecules without repeat blocks were predominantly composed of amino acids that exhibit a high β-sheet, β-turn, and/or random coil potential. Generally, within most of these molecules, the presence of repeat segments usually predicts the location of a helical domain. One of the possible pressures for the maintenance of repeat blocks is the preservation of the helix potential within specific regions of these molecules, the presence of which may determine an extended protein structure as has been shown for the M protein (Li, et al., 2013). An exception to this is found in the Sec10 protein, which is a predominantly helical molecule with limited repeat segments.
7-residue periodicity
Conformational analysis has revealed that most of the surface molecules that contain repeat sequences were found to be α-helical in those regions composed of repeats, while molecules without repeat blocks exhibited a high degree of β-sheet, β-turn, and random coil. However, except for M6 protein, little information is available to indicate that any of these surface proteins are also in a coiled-coil conformation. To answer this question, an algorithm was developed (Fischetti, Landau, Sellers, & Schmidt, 1993) and used to determine if a 7-residue periodicity also exists in those molecules that contain α-helical regions. When this algorithm was applied to representative sequences, extended regions of 7-residue periodicity were found in Arp4, FcRA, protein H, Pac, and spaP proteins. This strongly suggests that these molecules may attain a coiled-coil conformation within these α-helical segments. It is apparent that only those proteins that contained sequence repeats and exhibited high α-helical potential showed the presence of a 7-residue pattern of hydrophobic amino acids. On the other hand, even though the protein G molecule exhibited an extensive helix potential, the heptad pattern was only found in a limited region. Interestingly, however, the PAc protein from S. mutans, only has one small segment between residues 120 to 520 that displays repeat blocks, and this segment exhibits strong helix potential. This is precisely the region that was also found to contain a regular 7-residue heptad pattern. When taken together, these data strongly suggest that the coiled-coil conformation may also be a characteristic of surface proteins other than the M protein.
Size Variation
Size variation is a unique property of the M molecule. Using a broadly cross-reactive monoclonal antibody as a probe, the size of the M protein, which was extracted by solubilizing the cell wall from a number of streptococcal strains with a phage lysin, was examined by Western blot. M protein derived from 20 different serotypes exhibited variation in size ranging from 41 kDa to 80 kDa in molecular weight, depending on the strain (Fischetti, Jones, & Scott, 1985). Similar size variation was observed when streptococcal strains of the same serotype (M6) isolated over a period of 40 years were examined in a similar way. This variation in size may be explained by the observation that the M sequence contains extensive repeats both at the protein and DNA levels. Long, reiterated DNA sequences likely serve as substrates for recombinational events or for replicative slippage, generating deletions and duplications within the M protein gene and leading to the production of M proteins of different sizes.
Sequence analysis of the M6 gene isolates from local streptococcal outbreaks revealed that they are clonally related and are not the result of separate acquisitions of unrelated M6 strains during the course of the infection (Fischetti, Jarymowycz, Jones, & Scott, 1986). The observation that size variants occur among these clinical isolates suggests that the size mutant constituted the major organism in the streptococcal population at the time of isolation, and also suggests that a given size mutant has a selective advantage under clinical conditions. Perhaps serological pressure forces the appearance and maintenance of the size variant as a result of a local immune response.
Size variation is not only found in the M protein molecule but is also seen in other surface proteins on Gram-positive bacteria that also exhibit sequence repeats; for example, PspA from S. pneumoniae (Wanda & Curtiss, 1994), HagA from P. gingivalis (Kozarov, Whitlock, Dong, Carrasco, & Progulske-Fox, 1998), and protein A from S. aureus (Cheung & Fischetti, 1988).
Multifunctional Proteins
Of the numerous proteins identified on the surface of streptococci and other Gram-positive organisms, the great majority contain domains that bind molecules found in body secretions (Table 1), which include immunoglobulins (IgA and IgG), albumin, fibronectin (Yamaguchi, Terao, & Kawabata, 2013), and fibrinogen (Anderson, et al., 2014). Other proteins have also been identified, based on the presence of domains exhibiting certain enzymatic activity. From current data, it is clear that the majority of these surface proteins are multifunctional, where the identified function is limited to only one domain of the multi-domain protein. In several cases, the function of the other region/s has not been identified. For example, as described above, it appears that many of the surface proteins contain a repeat region close to the cell wall and an extended non-repetitive domain N-terminal to this (see Figure 7). In most cases, the binding domain (i.e., fibronectin or immunoglobulin) is localized to the repeat domain, while the N-terminal region exhibits no known function. In the case of streptococcal serum opacity factor, both domains have been defined (Rakonjac, Robbins, & Fischetti, 1995); the repeats bind fibronectin, while the N-terminal domain has the catalytic function that cleaves ApoA1 of high-density lipoprotein and results in serum opacity. In the case of highly repetitive proteins, such as M protein and protein G, each repeat domain has a specific function. In M protein, the A repeats bind albumin, the B repeats fibrinogen (Rýc, Beachey, & Whitnack, 1989; Horstmann, Sievertsen, Leippe, & Fischetti, 1992), and the C repeats factor H of complement (Horstmann, Sievertsen, Knobloch, & Fischetti, 1988) and keratinocytes (Okada, Liszewski, Atkinson, & Caparon, 1995). In protein G, one repeat domain binds IgG, while the second binds albumin. The multifunctional feature of surface proteins is not limited to molecules anchored via their C-terminal region; proteins bound through hydrophobic/charge interactions also exhibit these same characteristics (see below).
Thus, in general, surface proteins on streptococci and other Gram-positive organisms are multifunctional molecules with at least two (and in some cases, three or more) independent functions. Given the fact that several different proteins that contain multifunctional domains may be found on the cell surface of a single streptococcus, the complexity associated with the bacterial surface can become enormous.
Surface Proteins Anchored by Charge and/or Hydrophobic Interactions
While the great majority of proteins identified on the surface of Gram-positive bacteria anchor through their C-termini, a few have been recently identified to bind through less defined charge and/or hydrophobic interactions.
Surface glycolytic enzymes
An in-depth analysis of the individual proteins identified in a cell wall extract of S. pyogenes revealed that some proteins were composed of cytosolic enzymes that are normally found in the glycolytic pathway (Pancholi & Fischetti, 1997). A total of five enzymes have been identified (trios-phosphate isomerase, glyceraldehyde-3-phosphate dehydrogenase [GAPDH], phosphoglycerate kinase, phosphoglycerate mutase, and α-enolase), which form a short contiguous segment of the glycolytic pathway (Reiss, Kanety, & Schlessinger, 1986) (Figure 8). Interestingly, these enzymes form a complex involved in the production of ATP. All five of these enzymes have been identified on the surface of nearly all streptococcal groups, and certain ones have been identified on the surface of fungi, such as Candida albicans, and parasites, such as trypanosomes and schistosomes (Table 2). Thus, given the right substrate, whole streptococci have the capacity to produce ATP on their cell surface, increasing the complexity and potential of such bacterial surfaces.
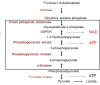
Figure 8.
A segment of the glycolytic pathway that depicts the enzymes located on the surface of the streptococcal cell wall (box) that produce NAD and ATP.
Table 2:
Enzymes identified on the surface of streptococcal groups, fungi, and parasites
Two surface glycolytic enzymes on the streptococcus have been the best characterized, GAPDH and α-enolase (Pancholi & Fischetti, 1993; Pancholi & Fischetti, 1998). Like other surface proteins on Gram-positives, GAPDH is a multifunctional protein with binding affinities for fibronectin and lysozyme, as well as cytoskeletal proteins, like actin and myosin. GAPDH has also been shown to have ADP-ribosylating activity in addition to its GAPDH activity. α-Enolase, is also multifunctional in its enzymatic activity and has the ability to specifically bind plasminogen, a plasma protease precursor (Pancholi & Fischetti, 1998). In some pathogens, only enolase is found on the surface. In these organisms, the surface enolase is used in the invasion of their host cells by binding plasminogen. Evidence shows that there is only one gene each for encoding GAPDH and enolase, and that both are produced in the cytosol and partially (30–40%) translocated to the cell surface (Derbise, Song, Parikh, Fischetti, & Pancholi, 2004). Since these molecules are not synthesized with an N-terminal signal sequence, it is unclear how they are ultimately translocated outside the bacteria. Also, because they do not contain an apparent anchor motif, the ways in which they are attached to the bacterial surface are also unknown. Since they can be removed with chaotropic agents, it is reasonable to assume that they are bound through charge and/or hydrophobic interactions.
Lipoproteins
Bacterial proteins may be lipid modified to facilitate their anchoring to the cytoplasmic membrane. Acylation effectively provides a membrane anchor allowing the now lipoprotein to function the aqueous environment. Bacterial lipoproteins have been shown to perform various roles, including nutrient uptake, signal transduction, adhesion, conjugation, and sporulation, and participate in antibiotic resistance, transport (such as ABC transporter systems) (Sutcliffe & Russell, 1995; Lampen & Nielsen, 1984). While a great deal is known about lipoproteins in Gram-negative bacteria, little is known about those from Gram-positive bacteria. In the case of pathogens, lipoproteins have been shown to play a direct role in virulence-associated functions, such as colonization, invasion, evasion of host defenses, and immunomodulation. Using a sequence-based analysis, Sutcliffe and Harrington examined the sequence of S. pyogenes for an N-terminal lipidation pattern G+LPP, and found that at least 25 probable lipoproteins may be present (Sutcliffe & Harrington, 2002). For example, lipoprotein FtsB in S. pyogenes is a component of the FtsABCD transporter that is responsible for ferrichrome binding and uptake (Li, et al., 2013). However, lipoproteins isolated from S. pyogenes after treatment with penicillin were complex, containing penicillin binding proteins, membrane proteins of the ExPortal (a membrane region responsible for assembly of secreted proteins), and hypothetical proteins (Biagini, et al., 2015).
Conclusion
Based on current information, the surface of the Gram-positive bacterial cell wall is highly complex and could even be considered a distinct organelle, since it is composed of proteins with specific binding functions, enzymatic activity, and the ability to generate energy. Considering the fact that there could be more than 25 different proteins (anchored in different ways) on the cell surface, each with the potential of up to three functional domains, suggests that more than 75 independent functions could potentially be present on the cell surface—much more than had ever been anticipated.
Because surface proteins on streptococci and other Gram-positives do not have cytoplasmic domains, it is unlikely that the binding of these molecules to specific ligands induces a signal in the microbe to activate a gene product. It is more likely that binding initiates a conformational signal on the cell surface to perform a specific function. For example, streptococci usually infect the pharynx (and particularly the tonsils) through contact with contaminated saliva. Upon entering the oral cavity, the organism first encounters the mucus, which coats the mucosal epithelium. Soluble components found in the mucus such as IgA, IgG, albumin, fibronectin, and others, are able to interact with their respective binding proteins on the streptococcal surface. Among other functions, this binding may initiate a set of conformational events on the surface of the bacterial particle to drive the organism through the mucus to the epithelial surface. The energy required for these processes may be derived from surface glycolytic enzymes necessary to generate ATP. This must all occur quickly or the organism will be swept into the gut and eliminated. Thus, the molecules necessary to initiate infection at all body sites are poised and ready on the cell surface.
References
- Agarwal B. L. Rheumatic heart disease unabated in developing countries. Lancet. 1981;2(8252):910–911. [PubMed: 6117691]
- Anderson E. L., Cole J. N., Olson J., Ryba B., Ghosh P., Nizet V. The fibrinogen-binding M1 protein reduces pharyngeal cell adherence and colonization phenotypes of M1T1 group A Streptococcus. The Journal of Biological Chemistry. 2014;289(6):3539–3546. [PMC free article: PMC3916555] [PubMed: 24356958]
- Beachey E. H., Campbell G. L., Ofek I. Peptic digestion of streptococcal M protein. II. Extraction of M antigen from group A streptococci with pepsin. Infection and Immunity. 1974;9(5):891–896. [PMC free article: PMC414902] [PubMed: 4132911]
- Beachey E. H., Seyer J. M., Dale J. B., Simpson W. A., Kang A. H. Type-specific protective immunity evoked by synthetic peptide of Streptococcus pyogenes M protein. Nature. 1981;292(5822):457–459. [PubMed: 6166868]
- Berge A., Sjöbring U. PAM, a novel plasminogen-binding protein from Streptococcus pyogenes. The Journal of Biological Chemistry. 1993;268(34):25417–25424. [PubMed: 8244975]
- Berry A. M., Lock R. A., Thomas S. M., Rajan D. P., Hansman D., Paton J. C. Cloning and nucleotide sequence of the Streptococcus pneumoniae hyaluronidase gene and purification of the enzyme from recombinant Escherichia coli. Infection and Immunity. 1994;62(3):1101–1108. [PMC free article: PMC186229] [PubMed: 8112843]
- Bessen D. E., Fischetti V. A. Nucleotide sequences of two adjacent M and M-like protein genes of group A streptococci: different RNA transcript levels and identification of a unique IgA-binding protein. Infection and Immunity. 1992;60(1):124–135. [PMC free article: PMC257512] [PubMed: 1370269]
- Biagini M., Garibaldi M., Aprea S., Pezzicoli A., Doro F., Becherelli M., et al. The Human Pathogen Streptococcus pyogenes Releases Lipoproteins as Lipoprotein-rich Membrane Vesicles. Molecular & Cellular Proteomics. 2015;14(8):2138–2149. [PMC free article: PMC4528243] [PubMed: 26018414]
- Breese, B. B. (1978). Beta hemolytic streptococcal diseases. Boston: Houghton Mifflin Professional Publishers.
- Burne R. A., Penders J. E. Characterization of the Streptococcus mutans GS-5 fruA gene encoding Exo-beta-D-fructosidase. Infection and Immunity. 1992;60(11):4621–4632. [PMC free article: PMC258211] [PubMed: 1398976]
- Cámara M., Boulnois G. J., Andrew P. W., Mitchell T. J. A neuraminidase from Streptococcus pneumoniae has the features of a surface protein. Infection and Immunity. 1994;62(9):3688–3695. [PMC free article: PMC303019] [PubMed: 8063384]
- Carapetis J. R., Steer A. C., Mulholland E. K., Weber M. The global burden of group A streptococcal diseases. The Lancet Infectious Diseases. 2005;5(11):685–694. [PubMed: 16253886]
- Carlsson F., Stålhammar-Carlemalm M., Flärdh K., Sandin C., Carlemalm E., Lindahl G. Signal sequence directs localized secretion of bacterial surface proteins. Nature. 2006;442:943–946. [PubMed: 16929299]
- Chen C. C., Cleary P. P. Complete nucleotide sequence of the streptococcal C5a peptidase gene of Streptococcus pyogenes. The Journal of Biological Chemistry. 1990;265(6):3161–3167. [PubMed: 2406246]
- Cheung A. L., Fischetti V. A. Variation in the expression of cell wall proteins of Staphylococcus aureus grown on solid and liquid media. Infection and Immunity. 1988;56(5):1061–1065. [PMC free article: PMC259762] [PubMed: 3281899]
- Cole R. M., Hahn J. J. Cell wall replication in Streptococcus pyogenes. Science. 1962;135(3505):722–724. [PubMed: 13880442]
- Coligan J. E., Kindt T. J., Krause R. M. Structure of the streptococcal groups A, A-variant and C carbohydrates. Immunochemistry. 1978;15(10-11):755–760. [PubMed: 85600]
- Coligan J. E., Schnute W. C., Kindt T. J. Immunochemical and Chemical Studies on Streptococcal Group-Specific Carbohydrates. The Journal of Immunology. 1975;114(6):1654–1658. [PubMed: 165237]
- Collins C. M., Kimura A., Bisno A. L. Group G streptococcal M protein exhibits structural features analogous to those of class I M protein of group A streptococci. Infection and Immunity. 1992;60(9):3689–3696. [PMC free article: PMC257378] [PubMed: 1500178]
- Cunningham M. W. Pathogenesis of group A streptococcal infections. Clinical Microbiology Reviews. 2000;13(3):470–511. [PMC free article: PMC88944] [PubMed: 10885988]
- Cunningham M. W., Beachey E. H. Peptic digestion of streptococcal M protein. I. Effect of digestion at suboptimal pH upon the biological and immunological properties of purified M protein extracts. Infection and Immunity. 1974;9(2):244–248. [PMC free article: PMC414793] [PubMed: 4205942]
- Dale J. B., Seyer J. M., Beachey E. H. Type-specific immunogenicity of a chemically synthesized peptide fragment of type 5 streptococcal M protein. The Journal of Experimental Medicine. 1983;158(5):1727–1732. [PMC free article: PMC2187117] [PubMed: 6195292]
- Derbise A., Song Y. P., Parikh S., Fischetti V. A., Pancholi V. Role of the C-terminal lysine residues of streptococcal surface enolase in Glu- and Lys-plasminogen-binding activities of group A streptococci. Infection and Immunity. 2004;72(1):94–105. [PMC free article: PMC343989] [PubMed: 14688086]
- Ferretti J. J., Russell R. R., Dao M. L. Sequence analysis of the wall-associated protein precursor of Streptococcus mutans antigen A. Molecular Microbiology. 1989;3(4):469–478. [PubMed: 2761387]
- Fischetti V. A. Streptococcal M protein: molecular design and biological behavior. Clinical Microbiology Reviews. 1989;2(3):285–314. [PMC free article: PMC358122] [PubMed: 2670192]
- Fischetti V. A., Jarymowycz M., Jones K. F., Scott J. R. Streptococcal M protein size mutants occur at high frequency within a single strain. The Journal of Experimental Medicine. 1986;164(4):971–980. [PMC free article: PMC2188411] [PubMed: 3760782]
- Fischetti V. A., Jones K. F., Scott J. R. Size variation of the M protein in group A streptococci. The Journal of Experimental Medicine. 1985;161(6):1384–1401. [PMC free article: PMC2187625] [PubMed: 2409199]
- Fischetti V. A., Landau G. M., Sellers P. H., Schmidt J. P. Identifying periodic occurrences of a template with applications to protein structure. Information Processing Letters. 1993;45(1):11–18.
- Fischetti V. A., Pancholi V., Schneewind O. Conservation of a hexapeptide sequence in the anchor region of surface proteins of Gram-positive cocci. Molecular Microbiology. 1990;4(9):1603–1605. [PubMed: 2287281]
- Fischetti V. A., Parry D. A., Trus B. L., Hollingshead S. K., Scott J. R., Manjula B. N. Conformational characteristics of the complete sequence of group A streptococcal M6 protein. Proteins. 1988;3(1):60–69. [PubMed: 3287371]
- Frithz E., Hedén L. O., Lindahl G. Extensive sequence homology between IgA receptor and M protein in Streptococcus pyogenes. Molecular Microbiology. 1989;3(8):1111–1119. [PubMed: 2691841]
- Galli D., Lottspeich F., Wirth R. Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1. Molecular Microbiology. 1990;4(6):895–904. [PubMed: 2120541]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. Journal of Molecular Biology. 1978;120(1):97–120. [PubMed: 642007]
- Gomi H., Hozumi T., Hattori S., Tagawa C., Kishimoto F., Björck L. The gene sequence and some properties of protein H. A novel IgG-binding protein. Journal of Immunology. 1990;144(10):4046–4052. [PubMed: 2332638]
- Haanes E. J., Cleary P. P. Identification of a divergent M protein gene and an M protein related gene family in serotype 49 Streptococcus pyogenes. Journal of Bacteriology. 1989;171(12):6397–6408. [PMC free article: PMC210527] [PubMed: 2687231]
- Hanski E., Caparon M. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(13):6172–6176. [PMC free article: PMC402144] [PubMed: 1385871]
- Heath D. G., Boyle M. D., Cleary P. P. Isolated DNA repeat region from fcrA76, the Fc-binding protein gene from an M-type 76 strain of group A streptococci, encodes a protein with Fc-binding activity. Molecular Microbiology. 1990;4(12):2071–2079. [PubMed: 2089220]
- Hedén L. O., Frithz E., Lindahl G. Molecular characterization of an IgA receptor from Group B Streptococci: sequence of the gene, identification of a proline-rich region with unique structure and isolation of N-terminal fragments with IgA-binding capacity. European Journal of Immunology. 1991;21(6):1481–1490. [PubMed: 2044657]
- Hollingshead S. K., Fischetti V. A., Scott J. R. Complete nucleotide sequence of type 6 M protein of the group A streptococcus: repetitive structure and membrane anchor. The Journal of Biological Chemistry. 1986;261(4):1677–1686. [PubMed: 3511046]
- Hollingshead S. K., Fischetti V. A., Scott J. R. Size variation in group A streptococcal M protein is generated by homologous recombination between intragenic repeats. Molecular and General Genetics. 1987;207(2-3):196–203. [PubMed: 3039291]
- Horstmann R. D., Sievertsen H. J., Knobloch J., Fischetti V. A. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein Factor H. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(5):1657–1661. [PMC free article: PMC279833] [PubMed: 2964038]
- Horstmann R. D., Sievertsen H. J., Leippe M., Fischetti V. A. Role of fibrinogen in complement inhibition by streptococcal M protein. Infection and Immunity. 1992;60(12):5036–5041. [PMC free article: PMC258274] [PubMed: 1452335]
- Jaffe J., Natanson-Yaron S., Caparon M. G., Hanski E. Protein F2, a novel fibronectin-binding protein from Streptococcus pyogenes, possesses two binding domains. Molecular Microbiology. 1996;21(2):373–384. [PubMed: 8858591]
- Jonsson H., Burtsoff-Asp C., Guss B. Streptococcal protein MAG - a protein with broad albumin binding specificity. Biochimica et Biophysica Acta. 1995;1249(1):65–71. [PubMed: 7766685]
- Kao S. M., Olmsted S. B., Viksnins A. S., Gallo J. C., Dunny G. M. Molecular and genetic analysis of a region of plasmid pCF10 containing positive control genes and structural genes encoding surface proteins involved in pheromone-inducible conjugation in Enterococcus faecalis. Journal of Bacteriology. 1991;173(23):7650–7664. [PMC free article: PMC212534] [PubMed: 1938961]
- Katsukawa C. Cloning and nucleotide sequence of type 3 M protein gene (emm3) consisting of an N-terminal variable portion and C-terminal conserved C repeat regions: relation to other genes of Streptococcus pyogenes. Kansenshogaku Zasshi. The Journal of the Japanese Association for Infectious Diseases. 1994;68(5):698–705. [PubMed: 8207299]
- Kelly C., Evans P., Bergmeier L., Lee S. F., Progulske-Fox A., Harris A. C., et al. Sequence analysis of a cloned streptococcal surface antigen I/II. FEBS Letters. 1989;258(1):127–132. [PubMed: 2687020]
- Kline J. B., Xu S., Bisno A. L., Collins C. M. Identification of a fibronectin-binding protein (GfbA) in pathogenic group G streptococci. Infection and Immunity. 1996;64(6):2122–2129. [PMC free article: PMC174045] [PubMed: 8675316]
- Kok J., Leenhouts K. J., Haandrikman A. J., Ledeboer A. M., Venema G. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Applied and Environmental Microbiology. 1988;54(1):231–238. [PMC free article: PMC202426] [PubMed: 3278687]
- Kozarov E., Whitlock J., Dong H., Carrasco E., Progulske-Fox A. The number of direct repeats in hagA is variable among Porphyromonas gingivalis strains. Infection and Immunity. 1998;66(10):4721–4725. [PMC free article: PMC108580] [PubMed: 9746569]
- Kreikemeyer B., Talay S. R., Chhatwal G. S. Characterization of a novel fibronectin-binding surface protein in group A streptococci. Molecular Microbiology. 1995;17(1):137–145. [PubMed: 7476200]
- Lampen J. O., Nielsen J. B. N-terminal glyceride-cysteine modification of membrane penicillinases in gram-positive bacteria. Methods in Enzymology. 1984;106:365–368. [PubMed: 6387378]
- Lancefield R. C. The antigenic complex of Streptococcus hemolyticus. I Demonstration of a type-specific substance in extracts of Streptococcus hemolyticus. The Journal of Experimental Medicine. 1928;47(1):91–103. [PMC free article: PMC2131344] [PubMed: 19869404]
- Lancefield R. C. Persistence of type-specific antibodies in man following infection with group A streptococci. The Journal of Experimental Medicine. 1959;110(2):271–292. [PMC free article: PMC2136986] [PubMed: 13673139]
- Lancefield R. C. Current knowledge of the type-specific M antigens of group A streptococci. The Journal of Immunology. 1962;89(3):307–313. [PubMed: 14461914]
- Li H., Li N., Xu Q., Xiao C., Wang H., Guo Z., et al. Lipoprotein FtsB in Streptococcus pyogenes binds ferrichrome in two steps with residues Tyr137 and Trp204 as critical ligands. PLoS One. 2013;8(6):e65682. [PMC free article: PMC3688767] [PubMed: 23840354]
- Lindgren P.-E., McGavin M., Signäs C., Guss B., Gurusiddappa S., Höök M., et al. Two different genes coding for fibronectin-binding proteins from Streptococcus dysgalactiae. The complete nucleotide sequences and characterization of the binding domains. European Journal of Biochemistry. 1993;214:819–827. [PubMed: 8319691]
- Manjula B. N., Khandke K. M., Fairwell T., Relf W. A., Sriprakash K. S. Heptad motifs within the distal subdomain of the coiled-coil rod region of M protein from rheumatic fever and nephritis associated serotypes of group A streptococci are distinct from each other: nucleotide sequence of the M57 gene... Journal of Protein Chemistry. 1991;10(4):369–384. [PubMed: 1781883]
- Marraffini L. A., Schneewind O. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Molecular Microbiology. 2006;62(5):1402–1417. [PubMed: 17074072]
- Maxted W. R. The indirect bactericidal test as a means of identifying antibody to the M antigen of Streptococcus pyogenes. The British Journal of Experimental Pathology. 1956;37(4):415–422. [PMC free article: PMC2082572] [PubMed: 13364152]
- Mazmanian S. K., Ton-That H., Schneewind O. Identification of sortase, the transpeptidase that anchors surface proteins to the cell wall envelope of Staphylococcus aureus. Molecular Microbiology. 2001a;40(5):1049–1057. [PubMed: 11401711]
- Mazmanian S. K., Ton-That H., Schneewind O. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Molecular Microbiology. 2001b;40(5):1049–1057. [PubMed: 11401711]
- McMillan D. J., Drèze P. A., Vu T., Bessen D. E., Guglielmini J., Steer A. C., et al. Updated model of group A Streptococcus M proteins based on a comprehensive worldwide study. Clinical Microbiology and Infection. 2013;19(5):E222–E229. [PMC free article: PMC4568957] [PubMed: 23464795]
- McNamara C., Zinkernagel A. S., Macheboeuf P., Cunningham M. W., Nizet V., Ghosh P. Coiled-coil irregularities and instabilities in Group A Streptococcus M1 are required for virulence. Science. 2008;319(5868):1405–1408. [PMC free article: PMC2288698] [PubMed: 18323455]
- Michel J. L., Madoff L. C., Olson K., Kling D. E., Kasper D. L., Ausubel F. M. Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(21):10060–10064. [PMC free article: PMC50277] [PubMed: 1438195]
- Miller L., Gray L., Beachey E., Kehoe M. Antigenic variation among group A streptococcal M proteins: Nucleotide sequence of the serotype 5 M protein gene and its relationship with genes encoding types 6 and 24 M proteins. The Journal of Biological Chemistry. 1988;263(12):5668–5673. [PubMed: 3281944]
- Mouw A. R., Beachey E. H., Burdett V. Molecular evolution of streptococcal M protein: Cloning and nucleotide sequence of type 24 M protein gene and relation to other genes of Streptococcus pyogenes. Journal of Bacteriology. 1988;170(2):676–684. [PMC free article: PMC210708] [PubMed: 3276665]
- Nilsson M., Frykberg L., Flock J. I., Pei L., Lindberg M., Guss B. A fibrinogen-binding protein of Staphylococcus epidermidis. Infection and Immunity. 1998;66(6):2666–2673. [PMC free article: PMC108254] [PubMed: 9596732]
- Okada N., Liszewski M. K., Atkinson J. P., Caparon M. Membrane cofactor protein (CD46) is a keratinocyte receptor for the M protein of group A streptococcus. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(7):2489–2493. [PMC free article: PMC42243] [PubMed: 7708671]
- Okahashi N., Sasakawa C., Yoshikawa M., Hamada S., Koga T. Molecular characterization of a surface protein antigen gene from serotype c Streptococcus mutans implicated in dental caries. Molecular Microbiology. 1989;3(5):673–678. [PubMed: 2761390]
- Olsson A., Eliasson M., Guss B., Nilsson B., Hellman U., Lindberg M., et al. Structure and evolution of the repetitive gene encoding streptococcal protein G. European Journal of Biochemistry. 1987;168(2):319–324. [PubMed: 3665928]
- O'Toole P. W., Stenberg L., Rissler M., Lindahl G. Two major classes in the M protein family in group A streptococci. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(18):8661–8665. [PMC free article: PMC49980] [PubMed: 1528877]
- Ozeri V., Rosenshine I., Ben-Ze'Ev A., Bokoch G. M., Jou T. S., Hanski E. De novo formation of focal complex-like structures in host cells by invading Streptococci. Molecular Microbiology. 2001;41(3):561–573. [PubMed: 11532125]
- Pancholi V., Fischetti V. A. Isolation and characterization of the cell-associated region of group A streptococcal M6 protein. Journal of Bacteriology. 1988;170(6):2618–2624. [PMC free article: PMC211179] [PubMed: 2453502]
- Pancholi V., Fischetti V. A. Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(17):8154–8158. [PMC free article: PMC47307] [PubMed: 8367477]
- Pancholi, V., & Fischetti, V. A. (1997). Identification of a glycolytic enzyme complex on the surface of group A streptococci. ASM General Meeting.
- Pancholi V., Fischetti V. A. α-Enolase, a Novel Strong Plasmin(ogen) Binding Protein on the Surface of Pathogenic Streptococci. The Journal of Biological Chemistry. 1998;273:14503–14515. [PubMed: 9603964]
- Perez-Casal J., Caparon M. G., Scott J. R. Introduction of the emm6 gene into an emm-deleted strain of Streptococcus pyogenes restores its ability to resist phagocytosis. Research in Microbiology. 1992;143(6):549–558. [PubMed: 1475516]
- Perry A. M., Ton-That H., Mazmanian S. K., Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. The Journal of Biological Chemistry. 2002;277:16241–16248. [PubMed: 11856734]
- Phillips G. N., Flicker P. F., Cohen C., Manjula B. N., Fischetti V. A. Streptococcal M protein: alpha-helical coiled-coil structure and arrangement on the cell surface. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(8):4689–4693. [PMC free article: PMC320228] [PubMed: 7029524]
- Rakonjac J. V., Robbins J. C., Fischetti V. A. DNA sequence of the serum opacity factor of group A streptococci: Identification of a fibronectin-binding repeat domain. Infection and Immunity. 1995;63(2):622–631. [PMC free article: PMC173041] [PubMed: 7822031]
- Rammelkamp C. H., Weaver R. S. Acute glomerulonephritis. The significance of the variation in the incidence of the disease. The Journal of Clinical Investigation. 1953;32(4):345–358. [PMC free article: PMC438348] [PubMed: 13052693]
- Raz A., Fischetti V. A. Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(47):18549–18554. [PMC free article: PMC2587614] [PubMed: 19017791]
- Raz A., Talay S. R., Fischetti V. A. Cellular aspects of the distinct M protein and SfbI anchoring pathways in Streptococcus pyogenes. Molecular Microbiology. 2012;84(4):631–647. [PMC free article: PMC3383811] [PubMed: 22512736]
- Reiss N., Kanety H., Schlessinger J. Five enzymes of the glycolytic pathway serve as substrates for purified epidermal-growth-factor-receptor kinase. The Biochemical Journal. 1986;239(3):691–697. [PMC free article: PMC1147341] [PubMed: 3030270]
- Robbins J. C., Spanier J. G., Jones S. J., Simpson W. J., Cleary P. P. Streptococcus pyogenes type 12 M protein regulation by upstream sequences. Journal of Bacteriology. 1987;169(12):5633–5640. [PMC free article: PMC214017] [PubMed: 2445730]
- Rocha C. L., Fischetti V. A. Identification and characterization of a novel fibronectin-binding protein on the surface of group A streptococci. Infection and Immunity. 1999;67(6):2720–2728. [PMC free article: PMC96575] [PubMed: 10338474]
- Rýc M., Beachey E. H., Whitnack E. Ultrastructural localization of the fibrinogen-binding domain of streptococcal M protein. Infection and Immunity. 1989;57(8):2397–2404. [PMC free article: PMC313460] [PubMed: 2473035]
- Schneewind O., Jones K. F., Fischetti V. A. Sequence and structural characterization of the trypsin-resistant T6 surface protein of group A streptococci. Journal of Bacteriology. 1990;172(6):3310–3317. [PMC free article: PMC209141] [PubMed: 2188957]
- Schneewind O., Mihaylova-Petkov D., Model P. Cell wall sorting signals in surface proteins of gram-positive bacteria. The EMBO Journal. 1993;12(12):4803–4811. [PMC free article: PMC413927] [PubMed: 8223489]
- Schneewind, O., Pancholi, V., & Fischetti, V. A. (1992). New Perspectives on Streptococci and Streptococcal Infections. In G. Orefici (Ed.), The role of the "LPSTGE" motif and C-terminal hydrophobic domain of streptococcal M6 protein on membrane anchoring (pp. 174-176). New York: Gustav Fischer Verlag.
- Scott J. R., Zähner D. Pili with strong attachments: Gram-positive bacteria do it differently. Molecular Microbiology. 2006;62(2):320–330. [PubMed: 16978260]
- Sela S., Aviv A., Tovi A., Burstein I., Caparon M. G., Hanski E. Protein F: an adhesin of Streptococcus pyogenes binds fibronectin via two distinct domains. Molecular Microbiology. 1993;10(5):1049–1055. [PubMed: 7934855]
- Sjöbring U. Isolation and molecular characterization of a novel albumin-binding protein from group G streptococci. Infection and Immunity. 1992;60(9):3601–3608. [PMC free article: PMC257367] [PubMed: 1500168]
- Smith H. E., Vecht U., Gielkens A. L., Smits M. A. Cloning and nucleotide sequence of the gene encoding the 136-kilodalton surface protein (muramidase-released protein) of Streptococcus suis type 2. Infection and Immunity. 1992;60(6):2361–2367. [PMC free article: PMC257166] [PubMed: 1587602]
- Sutcliffe I. C., Harrington D. J. Pattern searches for the identification of putative lipoprotein genes in Gram-positive bacterial genomes. Microbiology. 2002;148(Pt 7):2065–2077. [PubMed: 12101295]
- Sutcliffe I. C., Russell R. R. Lipoproteins of gram-positive bacteria. Journal of Bacteriology. 1995;177(5):1123–1128. [PMC free article: PMC176714] [PubMed: 7868582]
- Suvorov A. N., Flores A. E., Ferrieri P. Cloning of the glutamine synthetase gene from group B streptococci. Infection and Immunity. 1997;65(1):191–196. [PMC free article: PMC174575] [PubMed: 8975911]
- Swanson J., Hsu K. C., Gotschlich E. C. Electron microscopic studies on streptococci. I. M antigen. The Journal of Experimental Medicine. 1969;130(5):1063–1091. [PMC free article: PMC2180487] [PubMed: 5347694]
- Talay S. R., Valentin-Weigand P., Jerlström P. G., Timmis K. N., Chhatwal G. S. Fibronectin-binding protein of Streptococcus pyogenes: sequence of the binding domain involved in adherence of streptococci to epithelial cells. Infection and Immunity. 1992a;60(9):3837–3844. [PMC free article: PMC257397] [PubMed: 1386839]
- Talay S. R., Valentin-Weigand P., Jerlström P. G., Timmis K. N., Chhatwal G. S. Fibronectin-binding protein of Streptococcus pyogenes: sequence of the binding domain involved in adherence of streptococci to epithelial cells. Infection and Immunity. 1992b;60(9):3837–3844. [PMC free article: PMC257397] [PubMed: 1386839]
- Terao Y., Kawabata S., Kunitomo E., Murakami J., Nakagawa I., Hamada S. Fba, a novel fibronectin-binding protein from Streptococcus pyogenes, promotes bacterial entry into epithelial cells, and the fba gene is positively transcribed under the Mga regulator. Molecular Microbiology. 2001;42(1):75–86. [PubMed: 11679068]
- Timoney J. F., Artiushin S. C., Boschwitz J. S. Comparison of the sequences and functions of Streptococcus equi M-like proteins SeM and SzPSe. Infection and Immunity. 1997;65(9):3600–3605. [PMC free article: PMC175512] [PubMed: 9284125]
- Tokuda M., Okahashi N., Takahashi I., Nakai M., Nagaoka S., Kawagoe M., et al. Complete nucleotide sequence of the gene for a surface protein antigen of Streptococcus sobrinus. Infection and Immunity. 1991;59(9):3309–3312. [PMC free article: PMC258171] [PubMed: 1840575]
- Wanda S. Y., Curtiss R. Purification and characterization of Streptococcus sobrinus dextranase produced in recombinant Escherichia coli and sequence analysis of the dextranase gene. Journal of Bacteriology. 1994;176(13):3839–3850. [PMC free article: PMC205580] [PubMed: 8021165]
- Wannamaker L. W. The chains that link the throat to the heart. Circulation. 1973;48(1):9–18. [PubMed: 4592577]
- Yamaguchi M., Terao Y., Kawabata S. Pleiotropic virulence factor - Streptococcus pyogenes fibronectin-binding proteins. Cellular Microbiology. 2013;15(4):503–511. [PubMed: 23190012]
- An M protein coiled coil unfurls and exposes its hydrophobic core to capture LL-37.[Elife. 2022]An M protein coiled coil unfurls and exposes its hydrophobic core to capture LL-37.Kolesinski P, Wang KC, Hirose Y, Nizet V, Ghosh P. Elife. 2022 Jun 21; 11. Epub 2022 Jun 21.
- Review Adhesion and invasion of Streptococcus pyogenes into host cells and clinical relevance of intracellular streptococci.[Streptococcus pyogenes: Basic ...]Review Adhesion and invasion of Streptococcus pyogenes into host cells and clinical relevance of intracellular streptococci.Rohde M, Cleary PP. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. 2016
- Heptad motifs within the distal subdomain of the coiled-coil rod region of M protein from rheumatic fever and nephritis associated serotypes of group A streptococci are distinct from each other: nucleotide sequence of the M57 gene and relation of the deduced amino acid sequence to other M proteins.[J Protein Chem. 1991]Heptad motifs within the distal subdomain of the coiled-coil rod region of M protein from rheumatic fever and nephritis associated serotypes of group A streptococci are distinct from each other: nucleotide sequence of the M57 gene and relation of the deduced amino acid sequence to other M proteins.Manjula BN, Khandke KM, Fairwell T, Relf WA, Sriprakash KS. J Protein Chem. 1991 Aug; 10(4):369-84.
- Coiled-coil structure of group A streptococcal M proteins. Different temperature stability of class A and C proteins by hydrophobic-nonhydrophobic amino acid substitutions at heptad positions a and d.[Biochemistry. 1997]Coiled-coil structure of group A streptococcal M proteins. Different temperature stability of class A and C proteins by hydrophobic-nonhydrophobic amino acid substitutions at heptad positions a and d.Cedervall T, Johansson MU, Akerström B. Biochemistry. 1997 Apr 22; 36(16):4987-94.
- Review Streptococci and the complement system: interplay during infection, inflammation and autoimmunity.[FEBS Lett. 2020]Review Streptococci and the complement system: interplay during infection, inflammation and autoimmunity.Syed S, Viazmina L, Mager R, Meri S, Haapasalo K. FEBS Lett. 2020 Aug; 594(16):2570-2585. Epub 2020 Aug 19.
- M Protein and Other Surface Proteins on Streptococci - Streptococcus pyogenesM Protein and Other Surface Proteins on Streptococci - Streptococcus pyogenes
- Cell Wall and Surface Molecules: Capsule - Streptococcus pyogenesCell Wall and Surface Molecules: Capsule - Streptococcus pyogenes
- PREDICTED: Mus musculus leucine rich repeat containing 27 (Lrrc27), transcript v...PREDICTED: Mus musculus leucine rich repeat containing 27 (Lrrc27), transcript variant X10, mRNAgi|1720424551|ref|XM_006536267.4|Nucleotide
- PREDICTED: Rattus norvegicus neuronal guanine nucleotide exchange factor (Ngef),...PREDICTED: Rattus norvegicus neuronal guanine nucleotide exchange factor (Ngef), transcript variant X1, mRNAgi|2678969225|ref|XM_039083015.2|Nucleotide
- P-type Cu(+) transporter [Caenorhabditis elegans]P-type Cu(+) transporter [Caenorhabditis elegans]gi|392897154|ref|NP_001255202.1|Protein
Your browsing activity is empty.
Activity recording is turned off.
See more...