NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Allotey J, Snell KIE, Smuk M, et al. Validation and development of models using clinical, biochemical and ultrasound markers for predicting pre-eclampsia: an individual participant data meta-analysis. Southampton (UK): NIHR Journals Library; 2020 Dec. (Health Technology Assessment, No. 24.72.)

Validation and development of models using clinical, biochemical and ultrasound markers for predicting pre-eclampsia: an individual participant data meta-analysis.
Show detailsIn this chapter, we describe the results of developing new prediction models using the IPPIC data sets. The methods for this chapter are detailed in Chapter 3, Development and validation of pre-eclampsia prediction models. We aimed to develop 18 models, one for each combination of the three outcomes (any-onset, early-onset and late-onset pre-eclampsia), two trimesters (predictors measured at trimester 1 or at trimester 2) and three predictor sets (clinical characteristics only, clinical characteristics plus biochemical markers, and clinical characteristics plus ultrasound markers).
Summary of international data sets and predictor availability
A total of 78 data sets were included in the IPPIC project. As explained in Chapter 3, the available data sets did not record all of the variables of interest, or the same combination of variables as other data sets. The timing of measurements also differed (i.e. trimester 1, trimester 2 or both). Potential predictors deemed ‘important’ based on the clinical consensus (see Table 3) were ranked according to the mean of their scores (ranging from 1, not important, to 5, very important; Table 7).
TABLE 7
Ranked clinical characteristics as potential predictors and mean of scores from the clinical consensus group
None of the individual IPPIC data sets had all of the clinical characteristics of interest, and therefore some variables had to be excluded to form the model development sets. We first excluded variables that were not recorded in many data sets or for which few data were available across the data sets with that variable (removed if the proportion of events in data sets with the variable accounted for < 5% of events across all data sets). Protein–creatinine ratio (PCR), urine dipstick, family history of pre-eclampsia and previous small for gestational age fetus were all excluded. We also excluded multiple pregnancy as we aimed to develop models applicable to women with singleton pregnancies. Variables were then removed according to their ranking (lowest-ranking first) until we had a reasonable number of data sets for model development. Substance misuse, mode of conception, smoking and ethnicity were removed. Twelve data sets had all of the remaining clinical variables of interest, and these are summarised in Table 8.
TABLE 8
Patient characteristics in the 12 IPPIC data sets used for model development, using all available data for each variable (excluding missing observations)
Biochemical and ultrasound markers were recorded in very few data sets (see Table 7). To develop models including either biochemical or ultrasound markers in addition to clinical characteristics, only subsets of the 12 data sets could be used. Four data sets included biochemical markers and six data sets included ultrasound markers. Estimated fetal weight centile was excluded as a potential predictor as none of the data sets recorded this variable.
Missingness and multiple imputation
Data sets were included if they recorded either first-trimester or second-trimester measurements for the potential predictors of interest; therefore, many variables were systematically missing (not recorded or recorded for < 10% of individuals) in a data set and some were partially missing (missing for some individuals) in a data set. Table 9 summarises the missingness for variables and outcomes in the development data sets.
TABLE 9
Number and proportion of observations missing values for each variable in each data set included in model development
When checking convergence following imputation of systematically missing predictors (as described in Chapter 3, Development and validation of pre-eclampsia prediction models), values for first-trimester BMI were deemed poorly imputed for POUCH131 (i.e. convergence was not achieved despite the large burn-in, and extreme values were imputed), so the data set was excluded when developing models using first-trimester clinical characteristics. Three studies108,115,173 were excluded from model development using second-trimester clinical characteristics, owing to poor imputation of blood pressures and BMI.
For model development including biochemical markers, all four data sets were included for the first-trimester models, and POUCH131 was excluded for the second-trimester models. For model development including ultrasound markers, two data sets135,161 were excluded for first-trimester models, and three data sets108,135,149 were excluded for second-trimester models. The relevant imputation checking plots are provided in Appendix 15 along with an explanation of why these studies were excluded. A summary of the overall sample size and number of events contributing to model development for each combination of trimester of measurement (first or second) and predictor set (clinical characteristics, clinical characteristics plus biochemical markers, clinical characteristics plus ultrasound markers) is given in Table 10.
TABLE 10
Summary of sample size and number of events used for model development
Models including clinical characteristics only
The model development process was performed for data sets imputed with each form of BMI, namely BMI, ln(BMI) and BMI–2. The resulting models and performance statistics for all clinical characteristic models predicting any-onset, early-onset and late-onset pre-eclampsia are presented in Appendix 16. As an example of how the functional form was decided for BMI, let us consider the model including first-trimester clinical characteristics for any-onset pre-eclampsia. The model that included BMI rather than ln(BMI) or BMI–2 had an overall calibration slope closest to 1 (with least heterogeneity across data sets), calibration-in-the large closest to 0 (with least heterogeneity across data sets), and the C-statistic with least heterogeneity across data sets (C-statistic estimates were very similar across the three models). Therefore, the model with BMI was selected. This process was repeated for models developed using clinical characteristics for all three pre-eclampsia outcomes using first-trimester measurements for potential predictors, and then separately for models using second-trimester measurements.
A summary of the predictors retained for each model after variable selection is given in Table 11. For all but one model, BMI was best modelled linearly if it was retained. However, for early-onset pre-eclampsia (for which there are fewer events), first-trimester BMI was modelled non-linearly using (BMI/10)–2. The relationship between BMI and risk (log-odds) of early pre-eclampsia is shown in Figure 12. Autoimmune disease was only retained in the model including second-trimester predictors for early-onset pre-eclampsia. Nulliparity was dropped from the models for early-onset pre-eclampsia and diabetes was dropped from the first-trimester model for late-onset pre-eclampsia and all of the second-trimester models. DBP was not retained in the first-trimester models for any-onset or late-onset pre-eclampsia but was retained in the model for early-onset pre-eclampsia and all second-trimester models.
TABLE 11
Summary of clinical characteristics retained in the models for any-, early- and late-onset pre-eclampsia
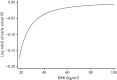
FIGURE 12
Relationship between first-trimester BMI and risk of early-onset pre-eclampsia when using (BMI/10)–2 transformation.
The parameter estimates for models with first-trimester predictors and second-trimester predictors are given in Tables 12 and 13, respectively. These are the developed models before adjustment for optimism due to overfitting. Increasing values of BMI, previous pre-eclampsia, history of hypertension, renal disease and diabetes were all associated with an increased risk of the pre-eclampsia outcomes. Increasing age was associated with a decrease in risk of pre-eclampsia. When retained in the models, increasing values of SBP and DBP were associated with an increase in risk for any-onset and late-onset pre-eclampsia, although second-trimester SBP was negatively associated with risk for early-onset pre-eclampsia.
TABLE 12
Parameter estimates for initial prediction models developed using first-trimester clinical characteristics to predict any-, early- or late-onset pre-eclampsia
TABLE 13
Parameter estimates for initial prediction models developed using second-trimester clinical characteristics to predict any-, early- or late-onset pre-eclampsia
Following internal validation, the predictive performance of the models is summarised in Table 14. The average (pooled) C-statistic for the models was close to 0.7, with considerable heterogeneity in the C-statistic across individual data sets. C-statistics were slightly higher for second-trimester models than for first-trimester models. The calibration slope was generally around 0.9 for models of any-onset and late-onset pre-eclampsia but was greater than 1 for early-onset pre-eclampsia (1.001 and 1.105). Again, there was large heterogeneity in the calibration slope across data sets for most models. Average calibration-in-the-large was close to zero but, again, there was large heterogeneity across data sets, suggesting that the baseline risk differs across the individual data sets and is not being fully captured by the predictors included in the models.
To illustrate the heterogeneity in predictive performance across individual data sets, Appendix 17 provides the forest plots of predictive performance measures for the second-trimester any-onset pre-eclampsia model. It is evident that there is large variability around the average values; for example, the observed calibration slope varies from 0.45 to 1.57 across data sets, and CIs often do not overlap.
Models including clinical characteristics and biochemical markers
Next, we examined whether or not biochemical markers should be included in the prediction models, in addition to the clinical characteristics identified for each outcome in the previous section (see Models including clinical characteristics only). Therefore, the clinical characteristics were forced into the models and only the biochemical markers were eligible for removal in the backwards elimination process. First-trimester BMI was previously modelled as (BMI/10)–2 for early pre-eclampsia; however, this transformation was not selected for any of the other clinical characteristic models and therefore may be a result of overfitting in data for which we have the fewest events. Therefore, for comparability and consistency across models, we used BMI rather than (BMI/10)–2 for early pre-eclampsia in the clinical plus biochemical marker models.
In the same way as for BMI in the clinical characteristics models, we considered non-normality for the biochemical markers by developing prediction models with biochemical markers on their original scale and compared this with models developed using natural logarithm-transformed biochemical marker values. Comparisons of biochemical marker and ln(biochemical marker) models are given in Appendix 18. For the first-trimester model for early pre-eclampsia, the model with better predictive performance came from the data with log-transformed biochemical markers; however, all biochemical markers were dropped from the model. Therefore, in this case, we selected the model that retained a biochemical marker in the model (see Table 16).
TABLE 16
Parameter estimates for initial prediction models using first-trimester clinical characteristics and biochemical markers to predict any-, early- or late-onset pre-eclampsia
Table 15 shows which biochemical markers were included in each model and if they were log-transformed. PAPP-A was not retained in any of the models, whereas sFlt-1 was retained in all first-trimester prediction models and the second-trimester model for early pre-eclampsia. PlGF was retained in all models except the first-trimester model for early pre-eclampsia. Models included the original biochemical marker values apart from the second-trimester model for early pre-eclampsia, which used ln(biochemical marker) values. For all but the second-trimester model for early pre-eclampsia, biochemical markers were negatively associated with the pre-eclampsia outcomes, so risk decreased with increasing biochemical marker values (Tables 16 and 17).
TABLE 15
Summary of biochemical markers retained in the models (alongside clinical characteristics) for any-, early- and late-onset pre-eclampsia using first- or second-trimester measurements
TABLE 17
Parameter estimates for initial prediction models using second-trimester clinical characteristics and biochemical markers to predict any-, early- or late-onset pre-eclampsia
Table 18 shows the average predictive performance for the models, obtained through internal validation using meta-analysis of the data set-specific performance statistics. Models with the clinical characteristics identified in Models including clinical characteristics only were also refitted in the same data so that it was possible to compare the predictive performance of models with and models without the biochemical markers in the same data sets. For all models, the average predictive performance improved with the addition of biochemical markers, and, for most models and performance statistics, heterogeneity across data sets was reduced (lower I2 and τ2 values). The average calibration slope was generally < 1 (between 0.857 and 0.961), except for the early-onset pre-eclampsia models, which had calibration slopes of 1.038 and 1.079 for the first- and second-trimester models, respectively.
Models including clinical characteristics and ultrasound markers
When considered in addition to clinical characteristics, ultrasound markers were not retained in any of the models fitted, whether using the original values or using the logarithm-transformed values. Therefore, the models reverted to the clinical characteristic models, which were reported in more detail and using more data sets in Models including clinical characteristics only.
Shrinkage and final models
Following model development and internal validation, shrinkage was applied to the beta coefficients and the final model equations are given in Table 19, along with the average performance statistics from meta-analysis across data sets for these models, including 95% CIs for the average performance and 95% prediction intervals for the performance of the model in a new but similar data set. Performance of the models in the individual data sets can be found in Appendix 19.
TABLE 19
Final model equations for each outcome, predictor type and trimester of measurement after shrinkage to adjust for optimism (overfitting)
After shrinkage and recalibration of the intercept, each model is, on average, perfectly calibrated across data sets. However, as was observed for existing models in Chapter 5, large heterogeneity remains in all of the performance statistics across data sets. The prediction intervals for potential performance in new settings are generally very wide. For example, the prediction interval for the calibration slope of model 4 ranges from 0.07 to 1.93. Therefore, although IPPIC models may predict well on average across populations, they may not be as accurate in particular populations. Figure 13 presents the calibration plots for model 1 (first-trimester clinical characteristics for the prediction of any pre-eclampsia) by data set. The model is fairly well calibrated for Baschat et al.;115 however, the predictions are too high for pregnant women in the WHO cohort,175 but not high enough for those at high risk in the POP cohort.161 Calibration plots for the other models (excluding for early pre-eclampsia, which had too few events in individual data sets) are given in Appendix 20. Heterogeneity in calibration performance could be reduced if, when applying the models in practice, model parameters (e.g. intercept) could be recalibrated to each population and setting. This would require local data for recalibration and model updating.
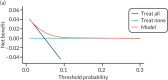
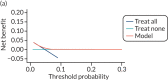
Decision curve analysis
Figures 14–17 are the decision curves in each data set for models predicting any pre-eclampsia. The decision curves show the net benefit or harm across different probability thresholds of the model and compared with the ‘treat-all’ and ‘treat-none’ strategies. Net benefit values are often best multiplied by 1000 to reveal the extra number of women who would be correctly treated per 1000 women who used the model, with none treated incorrectly.
Using first-trimester clinical characteristics (model 1; see Figure 14) shows some net benefit at thresholds around 0.05 in the Poston et al. 2015,149 Baschat et al.,115 STORK G,135 Vinter et al.173 and POP161 cohorts. The model shows little benefit or harm in SCOPE,42 Allen et al.,108 Antsaklis et al.,110 WHO175 and NICH LR.159 Using second-trimester clinical characteristics (model 4) shows some improvement in net benefit in SCOPE161 but little improvement in the other data sets (see Figure 15). In the data sets used for developing and validating models with first-trimester clinical characteristics and biochemical markers (model 7), there may be greater net benefit in POP161 but little improvement in the SCOPE42 or WHO175 cohorts (see Figure 16) compared with using clinical characteristics alone. Using second-trimester clinical characteristics and biochemical markers (model 10) shows a slight improvement in the SCOPE42 and POP161 cohorts (compared with first-trimester predictors), and the addition of second-trimester biochemical markers has slightly greater net benefit than in models including only second-trimester clinical characteristics (see Figure 17).
Decision curves for models predicting early-onset pre-eclampsia show no net benefit in most of the data sets, and decision curves for models predicting late-onset pre-eclampsia are similar to those for any-onset pre-eclampsia (see Appendix 21, Figures 38–45).
Summary
We used IPD from the IPPIC data sets to develop and validate new prediction models for early-onset, late-onset and any-onset pre-eclampsia using clinical characteristics alone or with the addition of biochemical markers. The IPPIC data sets used for model development and validation are heterogeneous, with different case mix in different data sets (e.g. owing to different inclusion and exclusion criteria). When the models were validated using an average intercept across data sets, the model performed better in some IPPIC data sets than in others even if the average performance was good. The same was observed in terms of net benefit in the individual data sets. In some data sets, there was potential for net benefit at certain thresholds, whereas there was very little or no net benefit when the models were applied in other data sets.
In summary, these prediction models have the potential to be useful in predicting pre-eclampsia in some populations; however, additional predictors may be needed, or the models may need to be tailored to improve the predictive performance across different settings and populations.
- Summary of international data sets and predictor availability
- Missingness and multiple imputation
- Models including clinical characteristics only
- Models including clinical characteristics and biochemical markers
- Models including clinical characteristics and ultrasound markers
- Shrinkage and final models
- Decision curve analysis
- Summary
- Development and validation of pre-eclampsia prediction models - Validation and d...Development and validation of pre-eclampsia prediction models - Validation and development of models using clinical, biochemical and ultrasound markers for predicting pre-eclampsia: an individual participant data meta-analysis
- Danio rerio hexosaminidase B (beta polypeptide) (hexb), transcript variant 2, mR...Danio rerio hexosaminidase B (beta polypeptide) (hexb), transcript variant 2, mRNAgi|836467586|ref|NM_001309863.1|Nucleotide
- Biomarkers for assessing acute kidney injury for people who are being considered...Biomarkers for assessing acute kidney injury for people who are being considered for admission to critical care: a systematic review and cost-effectiveness analysis
- Sec14l3 SEC14-like lipid binding 3 [Mus musculus]Sec14l3 SEC14-like lipid binding 3 [Mus musculus]Gene ID:380683Gene
Your browsing activity is empty.
Activity recording is turned off.
See more...