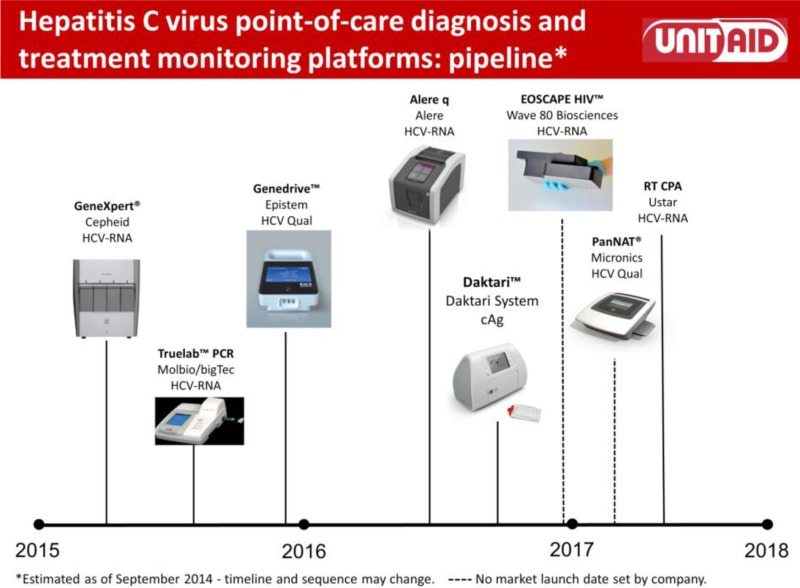
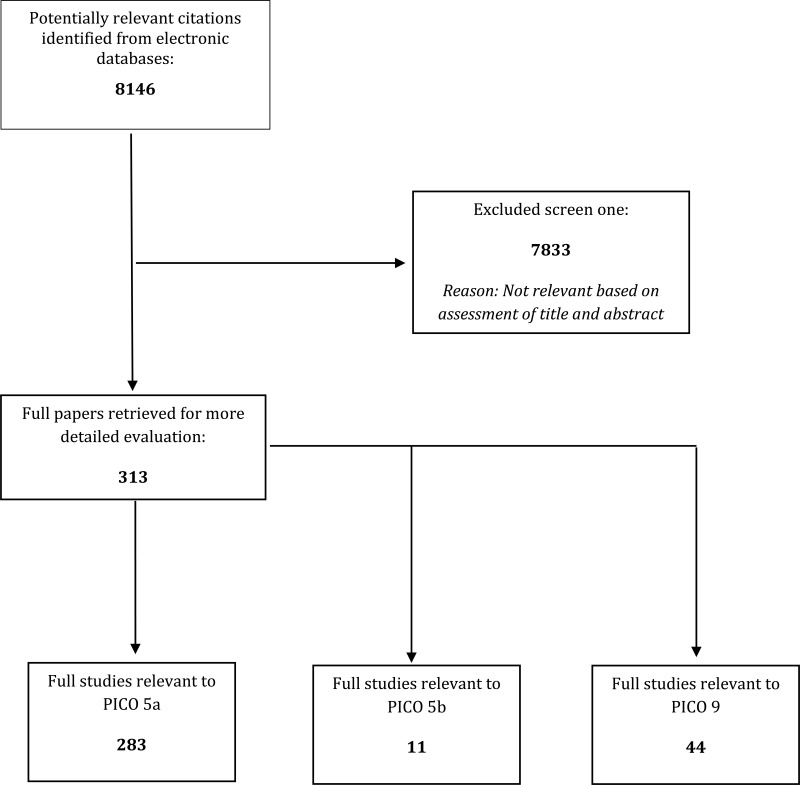
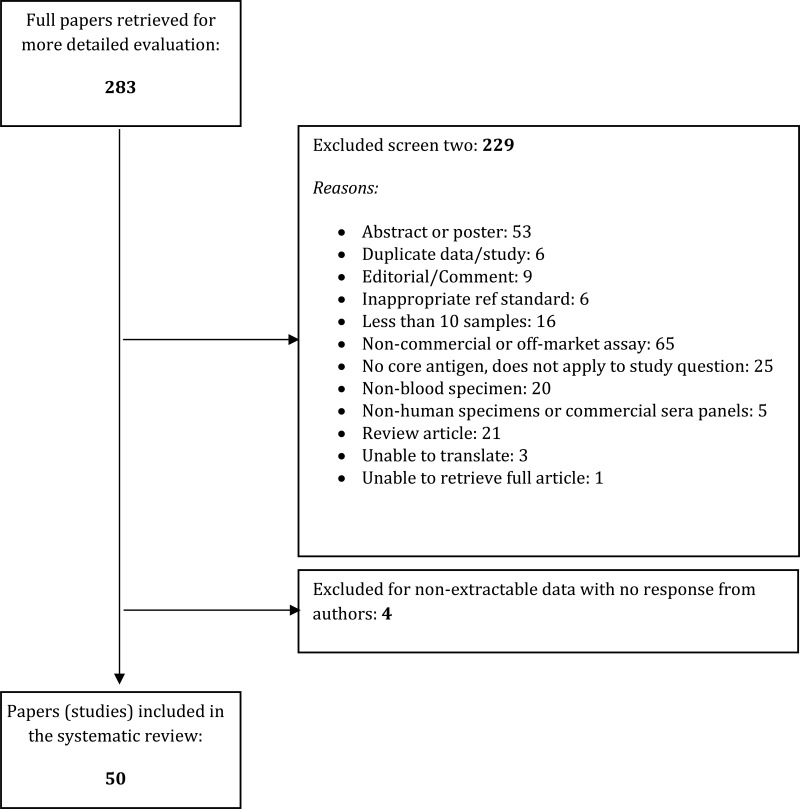
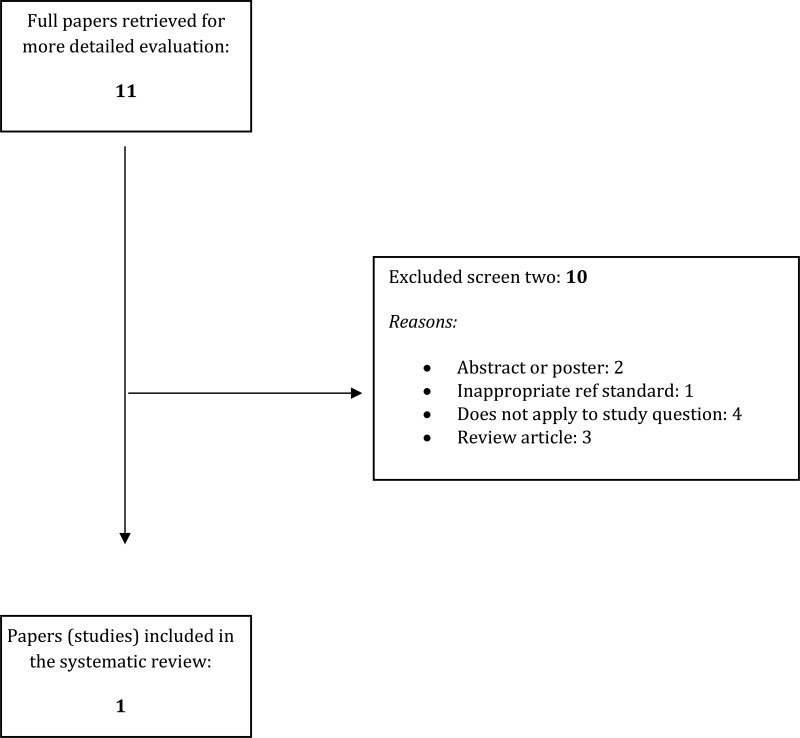
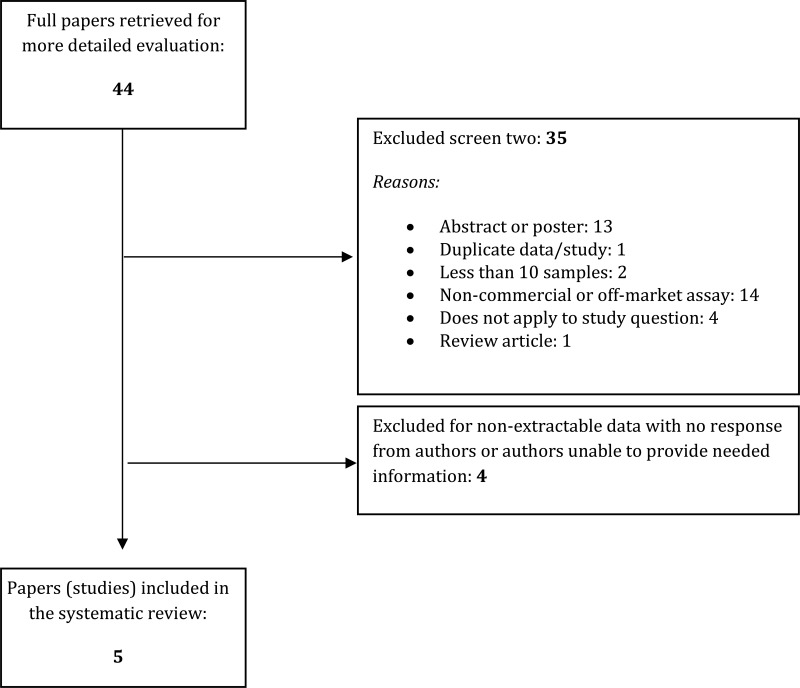
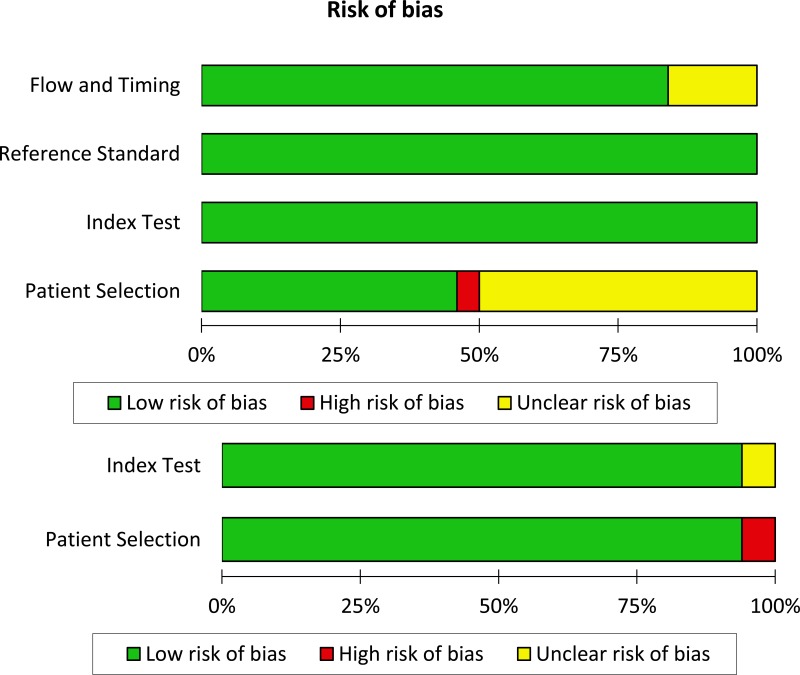
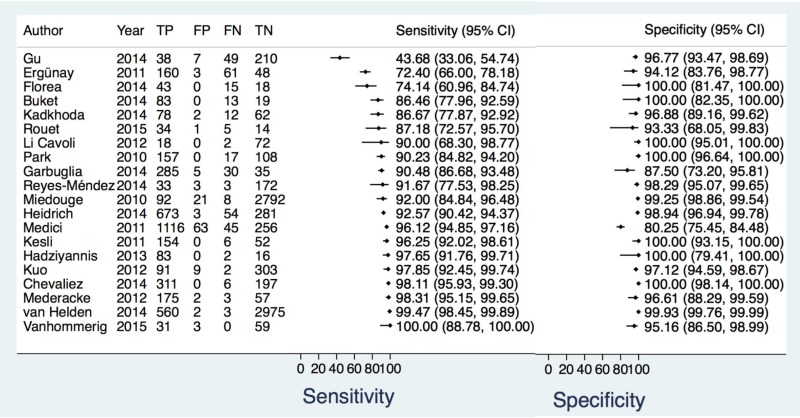
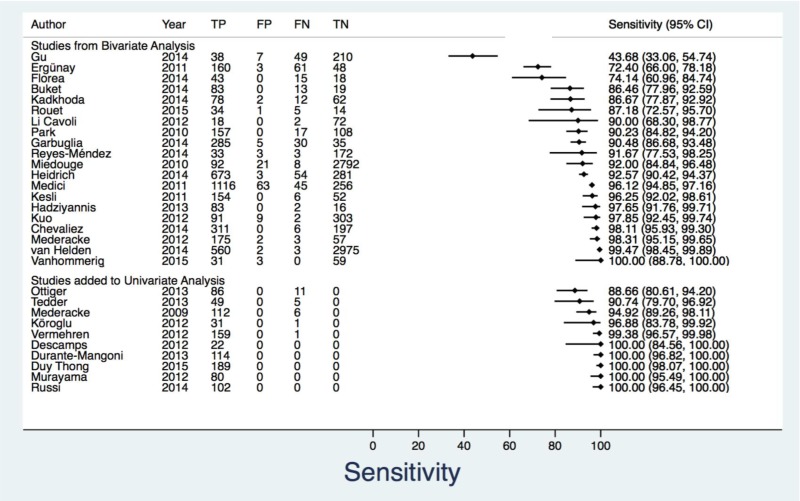
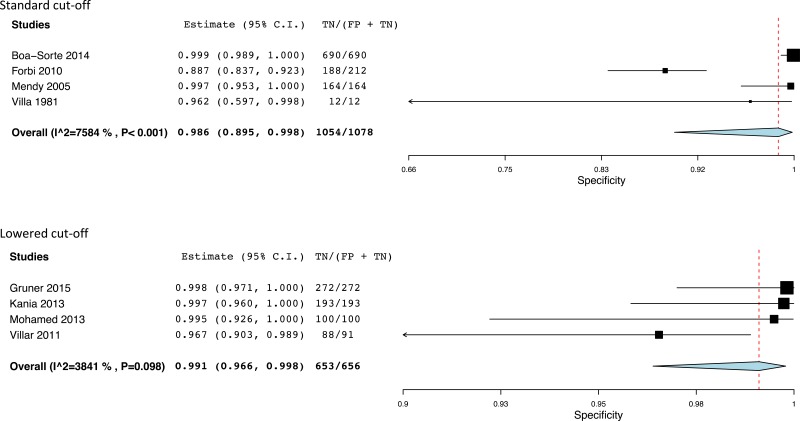
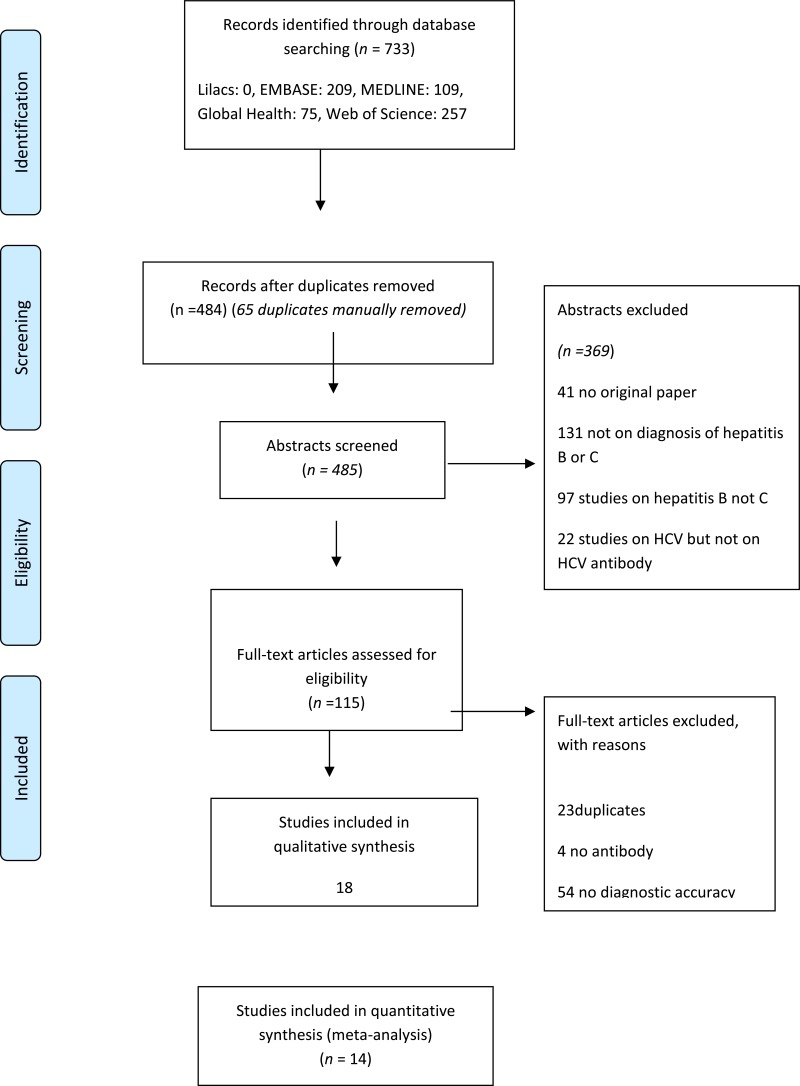
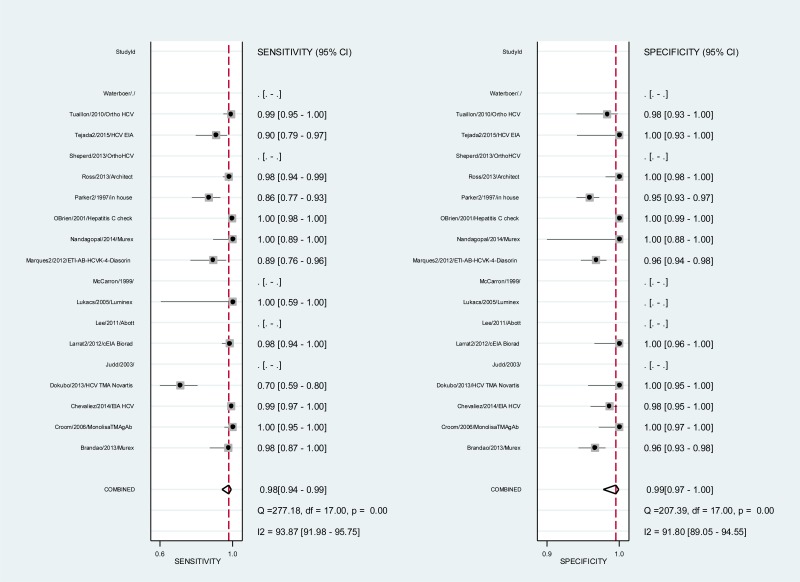
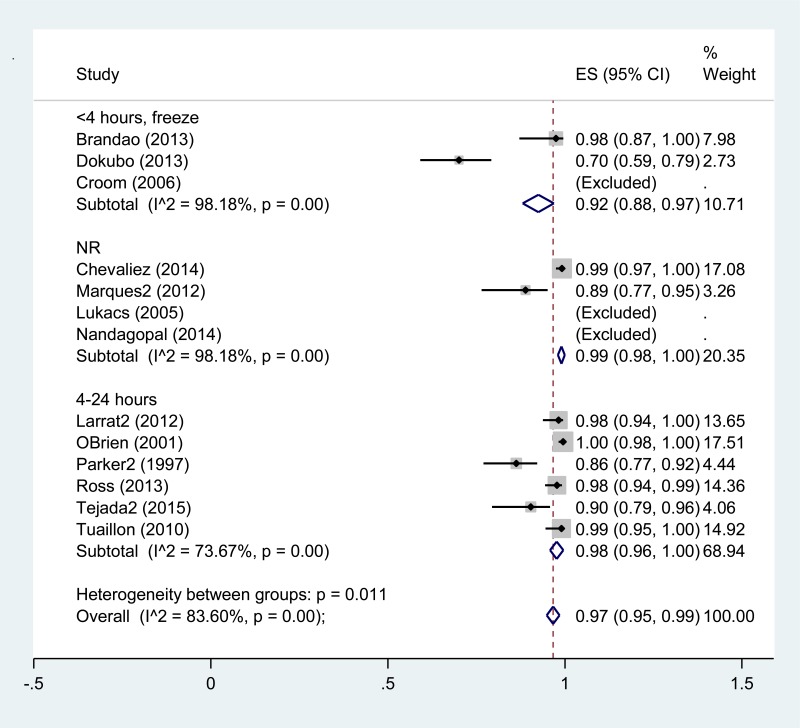
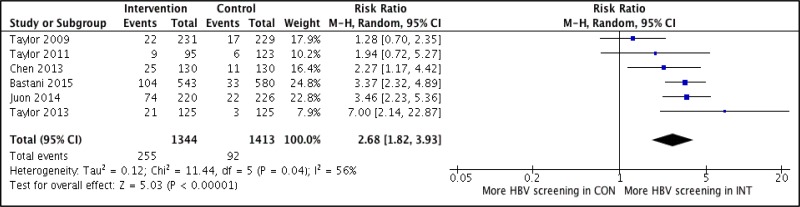
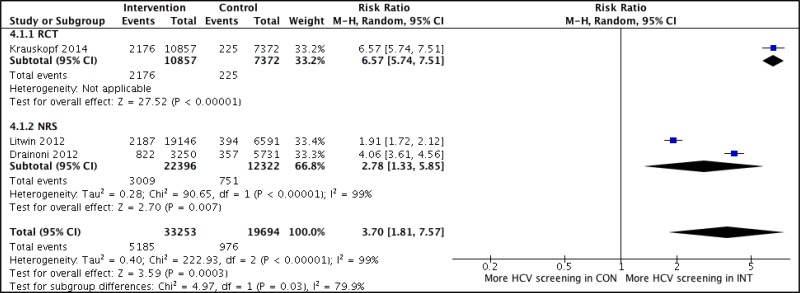
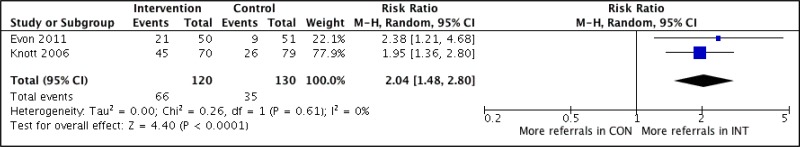
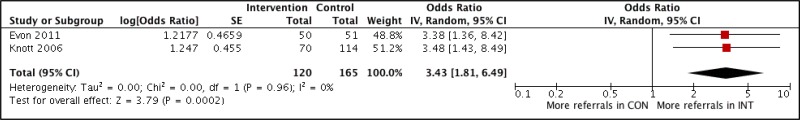
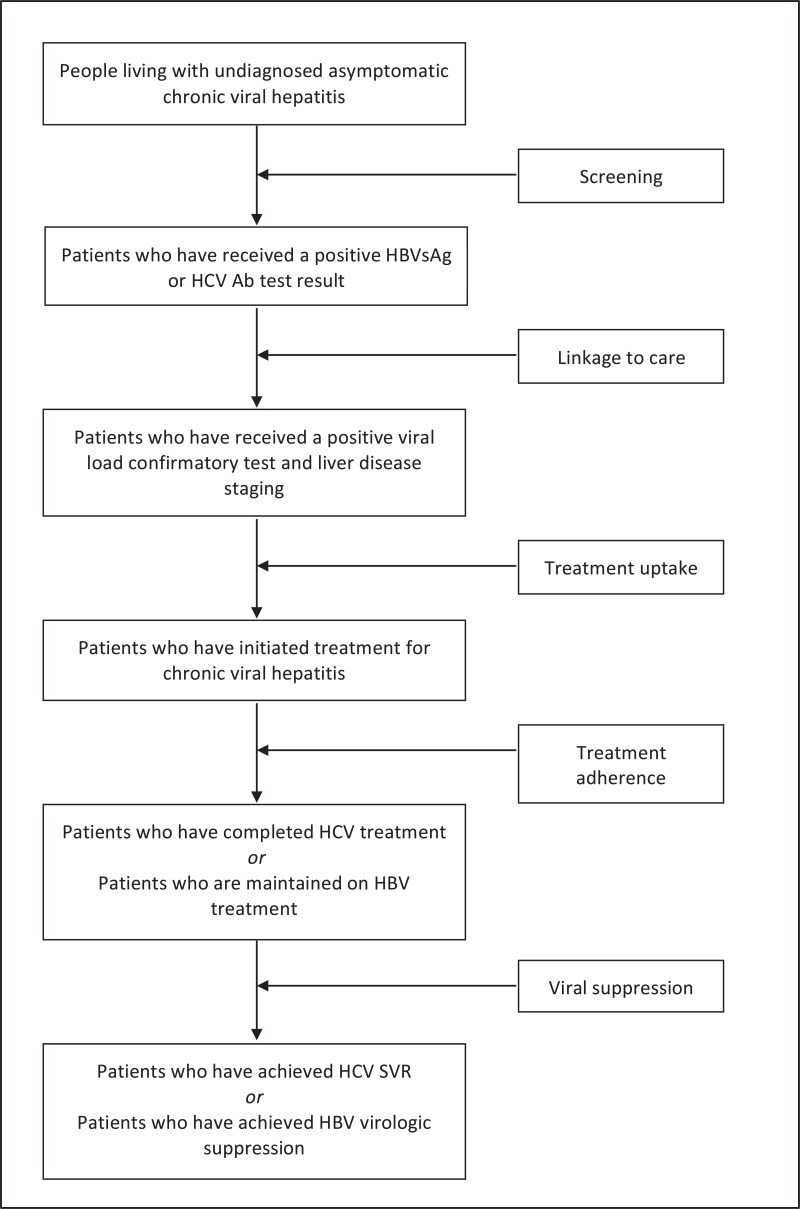
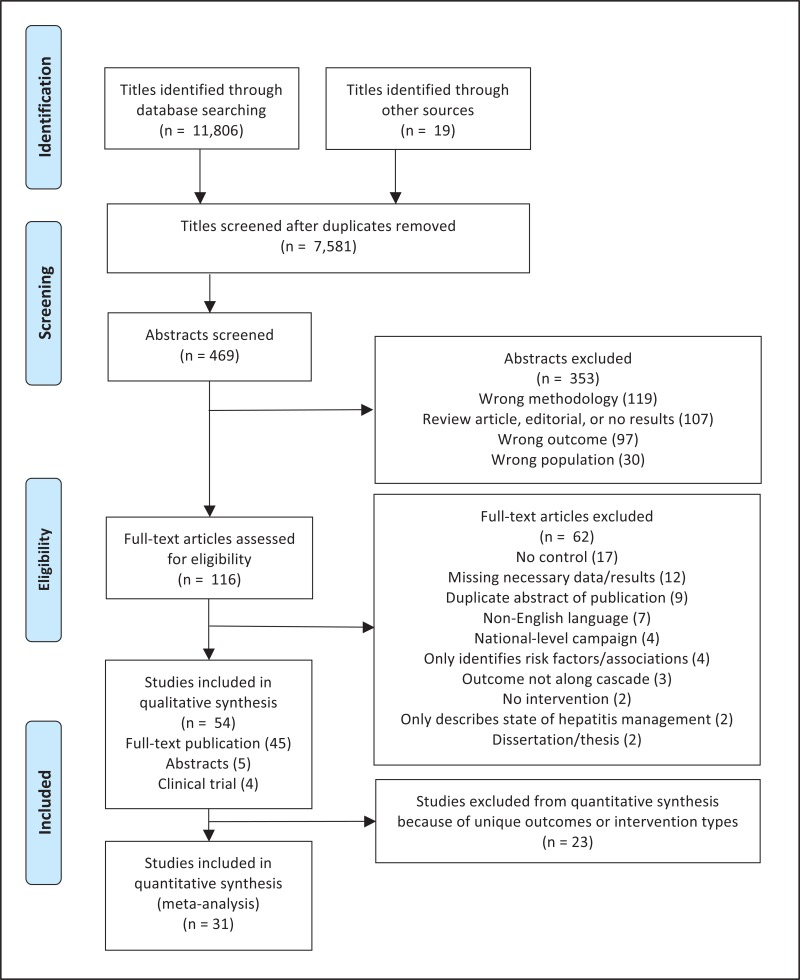
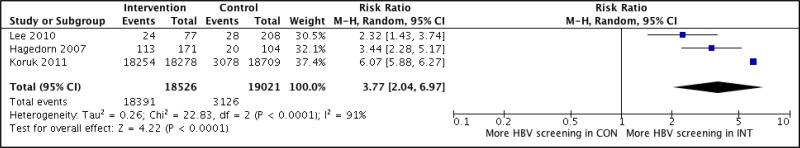
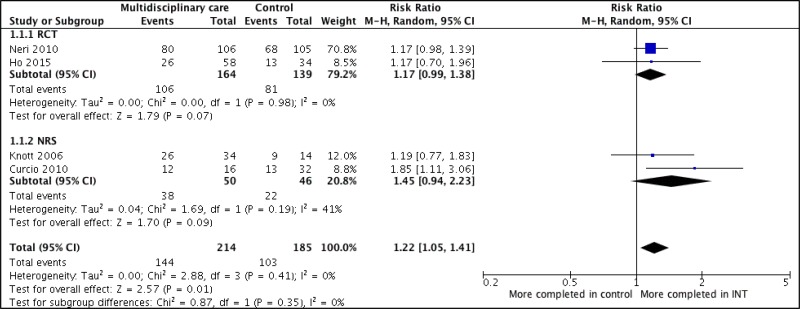
- 1.
Alados-Arboledas
JC
, Calbo-TorrecillasL, Lopez-PrietoMD, de Francisco-RamirezJL, de Miguel-SastreC. Clinical assessment of Monolisa HCV ag-ab ULTRA (Bio-Rad) in a general hospital. Enferm Infecc Microbiol Clin. 2007;25(3):172–6.
Inappropriate reference test [
PubMed: 17335695]
- 2.
Allain
JP
, CoghlanPJ, KenrickKG, WhitsonK, KellerA, CooperGJ, et al. Prediction of hepatitis C virus infectivity in seropositive Australian blood donors by supplemental immunoassays and detection of viral RNA. Blood. 1991;78(9):2462–8.
No HCV core antigen performed [
PubMed: 1657250]
- 3.
Alzahrani
AJ
. Analysis of hepatitis C virus core antigenemia in Saudi drug users. Saudi Med J. 2005;26(10):1645–6.
Editorial or comment [
PubMed: 16228075]
- 4.
Alzahrani
AJ
, ObeidOE, Al-AliA, ImamwardiB. Detection of hepatitis C virus and human immunodeficiency virus in expatriates in Saudi Arabia by antigen-antibody combination assays. J Infect Dev Ctries. 2009;3(3):235–8.
Less than 10 independent samples [
PubMed: 19759481]
- 5.
Aoyagi
K
, IidaK, OhueC, MatsunagaY, TanakaE, KiyosawaK, et al. Performance of a conventional enzyme immunoassay for hepatitis C virus core antigen in the early phases of hepatitis C infection. Clin Lab. 2001;47(3–4):119–27.
Non-commercial or off-market assay [
PubMed: 11294574]
- 6.
Aoyagi
K
, OhueC, IidaK, KimuraT, TanakaE, KiyosawaK, et al. Development of a simple and highly sensitive enzyme immunoassay for hepatitis C virus core antigen. J Clin Microbiol. 1999;37(6):1802–8.
Non-commercial or off-market assay [
PMC free article: PMC84955] [
PubMed: 10325327]
- 7.
Araujo
AC
, AstrakhantsevaIV, FieldsHA, KamiliS. Distinguishing acute from chronic hepatitis C virus (HCV) infection based on antibody reactivities to specific HCV structural and nonstructural proteins. J Clin Microbiol. 2011;49(1):54–7.
No HCV core antigen performed [
PMC free article: PMC3020409] [
PubMed: 21084519]
- 8.
Arrojo
IP
, ParejaMO, OrtaMDR, LuqueFN, LamasMCH, GordoFS, et al. Detection of a healthy carrier of HCV with no evidence of antibodies for over four years. Transfusion. 2003;43(7):953–7.
Less than 10 independent samples [
PubMed: 12823756]
- 9.
Attallah
AM
, IsmailH, TabllAA, ShibaGE, El-DosokyI. A novel antigen detection immunoassay for field diagnosis of hepatitis C virus infection. J Immunoassay Immunochem. 2003;24(4):395–407.
Non-commercial or off-market assay [
PubMed: 14677657]
- 10.
Attallah
AM
, OmranMM, NasifWA, GhalyMF, El-ShanshouryAERR, AbdallaMS, et al. Diagnostic performances of hepatitis C virus-NS4 antigen in patients with different liver pathologies. Arch Med Res. 2012;43(7):555–62.
Non-commercial or off-market assay [
PubMed: 23085447]
- 11.
Attallah
AM
, ShihaGE, MalakCAA, HagrasHE, Abdel-RazikWS, IsmailH. Utility of a novel HCVNS4 antigen detection immunoassay for monitoring treatment of HCV-infected individuals with pegylated interferon alpha-2a. Hepatol Res. 2004;28(2):68–72. Non-commercial or off-market assay
- 12.
Attia
MA
, ZekriAR, GoudsmitJ, BoomR, KhaledHM, MansourMT, et al. Diverse patterns of recognition of hepatitis C virus core and nonstructural antigens by antibodies present in Egyptian cancer patients and blood donors. J Clin Microbiol. 1996;34(11):2665–9.
No HCV core antigen performed [
PMC free article: PMC229382] [
PubMed: 8897161]
- 13.
Baggett
DW
, MoroneyS, SaewertM, JaczkoB, ZelechowskiJ, BahiC, et al. Dynamics of circulating HCV core antigen and HCV RNA in the early phase of HCV infection. Transfusion. 2000;40(10):26S. Abstract or poster
- 14.
Ballardini
G
, ManzinA, GiostraF, FrancesconiR, GroffP, GrassiA, et al. Quantitative liver parameters of HCV infection: relation to HCV genotypes, viremia and response to interferon treatment. J Hepatol. 1997;26(4):779–86.
Non-blood specimen [
PubMed: 9126789]
- 15.
Baranov
AV
, MaleevVV. Association between HCV RNA level and anti-HCV antibodies during chronic hepatitis C. Zh Mikrobiol Epidemiol Immunobiol. 2009(5):19–22.
No HCV Core antigen performed [
PubMed: 20063787]
- 16.
Bdour
S
. Hepatitis C virus infection in Jordanian haemodialysis units: serological diagnosis and genotyping. J Med Microbiol. 2002;51(8):700–4.
No HCV core antigen performed [
PubMed: 12171303]
- 17.
Beer
N
, ShinarE, NovackL, SafiJ, SolimanH, YaariA, et al. Accuracy of hepatitis C virus core antigen testing in pools among seroconverters. Transfusion. 2006;46(10):1822–8.
Noncommercial or off-market assay [
PubMed: 17002640]
- 18.
Berger
A
. Recent developments in hepatitis C infection (epidemiology, diagnosis and therapy). Laboratoriums Medizin. 2001;25(7–8):218–22. Review article
- 19.
Berger
A
, DoerrHW, PreiserW, WeberB. Lack of correlation between different hepatitis C virus screening and confirmatory assays. J Virol Methods. 1996;59(1–2):141–6.
No HCV core antigen performed [
PubMed: 8793841]
- 20.
Bochkova
G
, FominaS, PuzyrevV, ObriadinaA, BurkovA, UlanovaT. The evaluation of the ELISA kit nullDS-EIA-anti-HCVSPECTR-GMnull as supplemental assay for confirmation of anti-HCV screening positive results. Clin Microbiol Infect. 2011;17:S664. Abstract or poster
- 21.
Bochkova
G
, FominaS, PuzyrevV, ObriadinaA, BurkovA, UlanovaT. The evaluation of the new ELISA kit “EIA-anti- HCV-SPECTRUM-M” intended for separate detection of anti-IgM to different HCV antigens. J Viral Hepat. 2012;19:5–6. Abstract or poster
- 22.
Bouvier-Alias
M
, PatelK, DahariH, BeaucourtS, LarderieP, BlattL, et al. Clinical utility of total HCV core antigen quantification: a new indirect marker of HCV replication. Hepatology. 2002;36(1):211–8.
Non-commercial or off market assay [
PubMed: 12085367]
- 23.
Bouzgarrou
N
, FodhaI, BenOthman S, AchourA, GrattardF, TrabelsiA, et al. Evaluation of a total core antigen assay for the diagnosis of hepatitis C virus infection in hemodialysis patients. J Med Virol. 2005;77(4):502–8.
Non-commercial or off market assay [
PubMed: 16254976]
- 24.
Brandao
CPU
, MarquesBLC, MarquesVA, Villela-NogueiraCA, Do OKMR, de PaulaMT, et al. Simultaneous detection of hepatitis c virus antigen and antibodies in dried blood spots. J Clin Virol. 2013;57(2):98–102.
Inappropriate reference test [
PubMed: 23518440]
- 25.
Brody
RI
, EngS, MelamedJ, MizrachiH, SchneiderRJ, TobiasH, et al. Immunohistochemical detection of hepatitis C antigen by monoclonal antibody TORDJI-22 compared with PCR viral detection. Am J Clin Pathol. 1998;110(1):32–7.
Non-blood specimens [
PubMed: 9661920]
- 26.
Brojer
E
, GronowskaA, MedyńskaJ, GrabarczykP, MikulskaM, LȩtowskaM, et al. The hepatitis C virus genotype and subtype frequency in hepatitis C virus RNA-positive, hepatitis C virus antibody-negative blood donors identified in the nucleic acid test screening program in Poland. Transfusion. 2004;44(12):1706–10.
No HCV core antigen performed [
PubMed: 15584984]
- 27.
Brojer
E
, LiszewskiG, NiznikA, RosiekA, LetowskaM, PetersonJE, et al. Detection of HCV core antigen in HCV RNA positive, anti-HCV negative blood donations from Polish blood donors. Transfusion. 2001;41(2):304.
Editorial or comment [
PubMed: 11239242]
- 28.
Brojer
E
, GronowskaA, RosiekA, MikulskaM, LetowskaM. HCV core antigen detection and quantification of viremia in the “window period” of donors identified by routine HCV RNA screening in Poland. Vox Sang. 2005;89:92. Poster or abstract
- 29.
Burek
V
. Hepatitis C viral infection – news in diagnostics. Infektoloski Glasnik. 2002;22(1):27–9. Review article
- 30.
Busch
MP
, WrightDJ, HirschkornDF, BaggettD, MaretS, LeeSR, et al. Sensitivity of 1(st) and 2(nd) generation HCV antigen assays versus nucleic acid testing (NAT) for detection of ramp-up phase of HCV infection. Transfusion. 2001;41(9):3S. Abstract or poster
- 31.
Cagnon
L
, WagamanP, BartenschlagerR, PietschmannT, GaoTJ, KnetemanNM, et al. Application of the trak-C (TM) HCV core assay for monitoring antiviral activity in HCV replication systems. J Virol Methods. 2004;118(1):23–31.
Non-human subjects, non-commercial or off-market assay [
PubMed: 15158065]
- 32.
Cano
H
, CandelaMJ, LozanoML, VicenteV. Application of a new enzyme-linked immunosorbent assay for detection of total hepatitis C virus core antigen in blood donors. Transfus Med.. 2003;13(5):259–66.
Non-commercial or off-market assay [
PubMed: 14617336]
- 33.
Cao
H
, ZhangK, ShuX, LiG. The effect of hepatitis B virus infection on detection of hepatitis C virus core antigen. Hepatol Int2012;6(1):199. Abstract or poster
- 34.
Cao
H
, ZhangK, ShuX, XuQH, LiG. Detection of core antigen of hepatitis virus C in patients infected with hepatitis virus C and B. Zhonghua Gan Zang Bing Za Zhi. 2011;19(10):726–8.
Noncommercial or off-market assay [
PubMed: 22409841]
- 35.
Cao
H
, ZhangK, ShuX, XuQH, LiG. Detection of hepatitis C core antigen in intravenous drug addictions. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2011;25(4):304–6.
Noncommercial or off-market assay [
PubMed: 22097615]
- 36.
Carabaich
A
, RuvolettoM, BernardinelloE, TonoN, CavallettoL, ChemelloL, et al. Profiles of HCV core protein and viremia in chronic hepatitis C: possible protective role of core antigen in liver damage. J Med Virol. 2005;76(1):55–60.
Non-commercial or off market assay [
PubMed: 15778969]
- 37.
Carney
R
, MaranaoD, SudraR, ChaytorS, LabbettW, JohnsonM, et al. A hepatitis C virus core antigen assay is a cost-effective, sensitive and specific test in the detection of acute hepatitis C in HIV infected subjects. HIV Med. 2014;15:8. Abstract or poster
- 38.
Cavazza
S
, LaggingM. Indeterminate third-generation hepatitis C recombinant immunoblot assay and HCV RNA analysis: isolated reactivity against NS5 associated with HCV viraemia in clinical patients but not blood donors. Scand J Infect Dis. 2005;37(6–7):488–92.
No HCV core antigen performed [
PubMed: 16012009]
- 39.
Cetinić
Balent N
, MikulićR, ĐakovićRode O. Enzyme immunoassay for separate detection of anti-HCV antibodies to individual HCV antigens as a confirmatory assay in diagnostics of HCV infection. Infektoloski Glasnik. 2013;33(3):109–15. No HCV core antigen performed
- 40.
Chakravarti
A
, ChauhanMS, DograG, BanerjeeS. Hepatitis C virus core antigen assay: can we think beyond convention in resource limited settings? Braz J Infect Dis. 2013;17(3):369–74.
Non-commercial or off market assay [
PMC free article: PMC9427406] [
PubMed: 23602467]
- 41.
Challine
D
, DameronG, LapercheL, LarderieP, RigotP, ClaquinJ, et al. Does HCV core antigen or nucleic acid testing in graft donors improve organ transplantation viral safety? Transfusion. 2001;41(9):3S–4S. Abstract or poster
- 42.
- 43.
Choi
YJ
, HuY, GorisJ, KimE. Histopathology, viral antigen and RNA in hepatitis C virus-associated hepatitis. Lab Invest. 1997;76(1):803.
- 44.
Cividini
A
, CerinoA, MuzziA, FurioneM, RebucciC, SegagniL, et al. Kinetics and significance of serum hepatitis C virus core antigen in patients with acute hepatitis C. J Clin Microbiol. 2003;41(5):2144–6.
Less than 10 independent samples [
PMC free article: PMC154695] [
PubMed: 12734263]
- 45.
Courouce
AM
, BarinF, BotteC, LunelF, MaisonneuveP, ManiezM, et al. A comparative evaluation of the sensitivity of 7 anti-hepatitis-C virus screening tests. Vox Sang. 1995;69(3):213–6.
No HCV core antigen performed [
PubMed: 8578733]
- 46.
Couroucé
AM
, Le MarrecN, BouchardeauF, RazerA, ManiezM, LapercheS, et al. Efficacy of HCV core antigen detection during the preseroconversion period. Transfusion. 2000;40(10):1198–202.
Less than 10 independent samples [
PubMed: 11061855]
- 47.
Craxi
A
, ValenzaM, FabianoC, MagrinS, FiorentinoG, DiquattroO, et al. Third generation hepatitis C virus tests in asymptomatic anti-HCV-positive blood donors. J Hepatol. 1994;21(5):730–4.
No HCV core antigen performed [
PubMed: 7890886]
- 48.
Cresswell
F
, ShawS, HughesD, YoussefE, HomerG, Hassan-IbrahimM, et al. Hepatitis C antigen testing: a reliable alternative for diagnosing acute hepatitis C infection. HIV Med. 2014;15:137. Abstract or poster
- 49.
Cucchietti
A
, ParkerS, OldfieldL, LimasC, GarrodA, McBryneL. Seroconversion sensitivity of elisa screening assays for HCV, HIV and HBV antigens and/or antibodies. Vox Sang. 2011;101:87. Abstract or poster
- 50.
Daniel
HD
, VivekanandanP, RaghuramanS, SridharanG, ChandyGM, AbrahamP. Significance of the hepatitis C virus (HCV) core antigen as an alternative plasma marker of active HCV infection. Indian J Med Microbiol. 2007;25(1):37–42.
Non-commercial or off-market assay [
PubMed: 17377351]
- 51.
Daniel
HDJ
, ChandyGM, AbrahamP. Quantitation of hepatitis C virus using an in-house real-time reverse transcriptase polymerase chain reaction in plasma samples. Diagn Microbiol Infect Dis. 2008;61(4):415–20.
No HCV core antigen performed [
PubMed: 18486403]
- 52.
Dawson
G
. The potential role of HCV core antigen testing in diagnosing HCV infection. Antivir Ther. 2012;17(7 PART B):1431–5.
Review article [
PubMed: 23322590]
- 53.
Dawson
GJ
. HCV core antigen and combination (antigen/antibody) assays for the detection of early Seroconversion. J Med Virol. 2007;79:S54–S8. Review article
- 54.
Dawson
GJ
. HCV core antigen detection in seropositive samples. J Med Virol. 2007;79:S52–S3. Review article
- 55.
De Almeida
Ponde RA
. Enzyme-linked immunosorbent/chemiluminescence assays, recombinant immunoblot assays and nucleic acid tests in the diagnosis of HCV infection. Eur J Clin Microbiol. Infect Dis2013;32(8):985–8.
Review article [
PubMed: 23666504]
- 56.
Dickson
RC
, MizokamiM, OritoE, QianK, LauJYN. Quantification of serum HCV core antigen by a fluorescent enzyme immunoassay in liver transplant recipients with recurrent hepatitis C – clinical and virologic implications. Transplantation. 1999;68(10):1512–6.
Non-commercial or off-market assay [
PubMed: 10589948]
- 57.
Dow
BC
, MunroH, BuchananI, FergusonK, DavidsonF, LycettC, et al. Acute hepatitis C virus seroconversion in a Scottish blood donor: HCV antigen is not comparable with HCV nucleic acid amplification technology screening. Vox Sang. 2004;86(1):15–20.
Less than 10 independent samples [
PubMed: 14984555]
- 58.
Dubrous
P
, HugardL, TerrierF. Screening for HCV core antigen among HIV-positive patients infected by sexual transmission. Med Mal Infect. 2004;34(5):236–8.
Less than 10 independent samples [
PubMed: 16235605]
- 59.
Durante-Mangoni
E
, VallefuocoL, PernaE, IossaD, AndiniR, CaianielloC, et al. Correlates and prognostic value of hepatitis C virus core antigen serum levels during chronic infection and treatment. Clin Microbiol Infect. 2010;16:S317. Abstract or poster
- 60.
Dwyer
E
, RileyP, PakianathanM. Analysis of hepatitis C antigen testing in an urban sexual health clinic. HIV Med. 2014;15:111–2. Abstract or poster
- 61.
Eiras
A
, FrancoE, MontoroJA, PlanellesD, VillaescusaR. HCV NAT (minipool RT-PCR) and HCV core antigen ELISA. Transfusion. 2003;43(1):118; author reply -9.
Editorial or comment [
PubMed: 12519441]
- 62.
El Ekiaby
M
, LapercheS, MoftahF, BurnoufT, LelieN. The impact of different HCV blood screening technologies on the reduction of transfusion transmitted HCV infection risk in Egypt. Vox Sang. 2009;96:23–4. Abstract or poster
- 63.
Ellethy
AT
, SliemHA, HassanGMA. Updated molecular diagnosis of chronic hepatitis C. J Gastroenterol Hepatol Res. 2012;1(8):147–52. Review article
- 64.
Ergunay
K
, SenerB, AlpA, KarakayaJ, HascelikG. Quantitative determination of HCV core antigen: A marker for monitoring hepatitis C viraemia. Clin Microbiol Infect. 2011;17:S663. Abstract or poster
- 65.
Fabrizi
F
, LunghiG, AucellaF, ManganoS, BarbisoniF, BisegnaS, et al. Novel assay using total hepatitis C virus (HCV) core antigen quantification for diagnosis of HCV infection in dialysis patients. J Clin Microbiol. 2005;43(1):414–20.
Non-commercial or off-market assay [
PMC free article: PMC540167] [
PubMed: 15635003]
- 66.
Frank
K
, KarlA. Comparison of different confirmation test methods to anti-HCV used for repeatedly HCV-reactive samples. Infusionsther Transfusionsmed. 2001;28(Suppl. 1):47. No HCV core antigen performed
- 67.
Galli
C
. New strategies for the identification of active hepatitis C virus (HCV) infections. Biochim Clin. 2013;37:S137. Abstract or poster
- 68.
Galli
C
. New strategies for the screening of hepatitis C virus infection. Vox Sang. 2013;105:181. Abstract or poster
- 69.
Gaudy
C
, ThevenasC, TichetJ, MariotteN, GoudeauA, DuboisF. Usefulness of the hepatitis C virus core antigen assay for screening of a population undergoing routine medical checkup. J Clin Microbiol. 2005;43(4):1722–6.
Non-commercial or off-market assay [
PMC free article: PMC1081371] [
PubMed: 15814991]
- 70.
Gavarro
A
, CebolleroA, MartinezS, VeraguasA, AcostaA. Evaluation of a new method to determine core antigen of hepatitis C virus and its comparison with the determination of RNAHCV. Clin Chem Lab Med. 2011;49:S522–S. Abstract or poster
- 71.
Grant
PR
, SimsCM, TedderRS. Quantification of HCV RNA levels and detection of core antigen in donations before seroconversion. Transfusion. 2002;42(8):1032–6.
Non-commercial or off-market assay [
PubMed: 12385415]
- 72.
Gu
SJ
, LiuJ, ZhangHJ, GuBL, LaiHJ, ZhouHL, et al. Core antigen tests for hepatitis C virus: a meta-analysis. Mol Biol Rep. 2012;39(8):8197–208.
Review article [
PubMed: 22544611]
- 73.
Hadziyannis
E
, VassilopoulosD, GeorgiouA, SpanouF, KoskinasJ. Comparison of a new HCV core antigen automated immunoassay to quantitative HCV RNA measurement in patients with chronic hepatitis C. Hepatology. 2010;52(4):1230A. Abstract or poster
- 74.
Hayashi
K
, HasuikeS, KusumotoK, IdoA, UtoH, KenjiN, et al. Usefulness of a new immunoradiometric assay to detect hepatitis C core antigen in a community-based population. J Viral Hepat. 2005;12(1):106–10.
Non-commercial or off-market assay [
PubMed: 15655057]
- 75.
Heidrich
B
, PischkeS, KirschnerJ, SchneiderJ, MederackeI, RaupachR, et al. Performance of HCV-core antigen testing in liver transplanted patients. J Hepatol. 2011;54:S223. Abstract or poster
- 76.
Higashimoto
M
, TakahashiM, JokyuR, SyundouH, SaitoH. Improvement of sensitivity in the second generation HCV core antigen assay by a novel concentration method using polyethylene glycol (PEG). Rinsho Byori. 2007;55(11):1008–14.
Less than 10 independent samples [
PubMed: 18154032]
- 77.
Hmaïed
F
, BenMamou M, ArroujiZ, SlimA, BenRedjeb S. Use of combined detection of hepatitis C virus core antigen and antibodies to reduce the serological window-phase. Pathol Biol (Paris). 2007;55(2):121–6.
Inappropriate reference test [
PubMed: 16631320]
- 78.
Hosseini-Moghaddam
SM
, Iran-PourE, RotsteinC, HusainS, LillyL, RennerE, et al. Hepatitis C core Ag and its clinical applicability: potential advantages and disadvantages for diagnosis and follow-up? Rev Med Virol. 2012;22(3):156–65.
Review article [
PubMed: 22121001]
- 79.
Icardi
G
, AnsaldiF, BruzzoneBM, DurandoP, LeeS, de LuigiC, et al. Novel approach to reduce the hepatitis C virus (HCV) window period: clinical evaluation of a new enzyme-linked immunosorbent assay for HCV core antigen. J Clin Microbiol. 2001;39(9):3110–14.
Less than 10 independent samples [
PMC free article: PMC88305] [
PubMed: 11526137]
- 80.
Icardi
G
, BruzzoneB, GotaF, TorreF, GianniniE, MassoneL, et al. A new assay for hepatitis C virus (HCV) core antigen detection: an alternative to nucleic acid technologies in positive or indeterminate anti-HCV subjects?Ann Ig. 2003;15(6):863–70.
Non-commercial or off market assay [
PubMed: 15049543]
- 81.
Irshad
M
, DharI, JoshiYK. Significance of hepatitis C virus core protein in the diagnosis of hepatitis C virus infection in different liver diseases. J Investig Med. 2006;54(8):478–83.
Duplicate data [
PubMed: 17169273]
- 82.
Irshad
M
, DharI, Khushboo, SinghS, KapoorS. Comparison of serological and nucleic Acid based assays used to diagnose hepatitis C virus (HCV) infection in acute and chronic liver diseases. Int J Health Sci (Qassim). 2007;1(1):3–10.
No extractable data, no response from author [
PMC free article: PMC3068650] [
PubMed: 21475446]
- 83.
Ivanov
YD
, KayshevaAL, FrantsuzovPA, PleshakovaTO, KrohinNV, IzotovAA, et al. Detection of hepatitis C virus core protein in serum by atomic force microscopy combined with mass spectrometry. Int J Nanomedicine. 2015;10:1597–608.
Non-commercial or off market assay [
PMC free article: PMC4346358] [
PubMed: 25759582]
- 84.
Ivanyi-Nagy
R
, MakowskaZ, LastraML, DarlixJL. Overview on the properties and functions of the core protein of hepatitis C cvirus (HCV). In: BernhardtLV, editor. Nova Science Publishers; 2010:1–47. Textbook chapter
- 85.
Kamili
S
, DrobeniucJ, AraujoAC, HaydenTM. Laboratory diagnostics for hepatitis C virus infection. Clin Infect Dis. 2012;55:S43–S48.
Review article [
PubMed: 22715213]
- 86.
Kashiwakuma
T
, HasegawaA, KajitaT, TakataA, MoriH, OhtaY, et al. Detection of hepatitis C virus specific core protein in serum of patients by a sensitive fluorescence enzyme immunoassay (FEIA). J Immunol Methods. 1996;190(1):79–89.
Non-commercial or off-market assay [
PubMed: 8601714]
- 87.
Kenfe
FR
, UrbaczekAC, SilvaJC, NeóTA, Da SilvaFH, Da CostaPI. Development of diagnostic methods and study of the immunoreactivity of a mixture of recombinant core and E2 proteins fused to GST with control serum positive for hepatitis C. Talanta. 2013;110:32–8.
Noncommercial or off-market assay [
PubMed: 23618172]
- 88.
Kesli
R
, PolatH, TerziY, KurtogluMG, UyarY. Comparison of a newly developed automated and quantitative hepatitis C virus core antigen test with the hepatitis C virus RNA assay for the clinical usefulness of confirming anti-hepatitis C virus results. Clin Microbiol Infect. 2012;18:676. [
PMC free article: PMC3233016] [
PubMed: 21940466]
- 89.
Kocazeybek
B
, YukselP, SaribasS, CaliskanR, ErginS, AslanM, et al. New approaches in in-vitro diagnosis of hepatitis C infections: the diagnostic performance of new hepatitis C virus core antigen detection test. Int J Infect Dis. 2010;14:e228. Abstract or poster
- 90.
Krajden
M
, ShivjiR, GunadasaK, MakA, McNabbG, FriesenhahnM, et al. Evaluation of the core antigen assay as a second-line supplemental test for diagnosis of active hepatitis C virus infection. J Clin Microbiol. 2004;42(9):4054–9.
Non-commercial or off-market assay [
PMC free article: PMC516310] [
PubMed: 15364989]
- 91.
Kuo
YH
, LuSN. Is HCV core antigen (HCV Ag) an adequate marker for community screening? Hepatol Int. 2012;6(1):201–2. Abstract or poster
- 92.
Kups
J
, Wozniakowska-GesickaT. Evaluation of specific humoral response to hepatitis C core antigen in children. Przegl Lek. 2003;60(12):802–5.
No HCV core antigen performed [
PubMed: 15058020]
- 93.
Kurtz
JB
, BoxallE, QusirN, ShirleyJ, ColemanD, ChandlerC. The diagnostic significance of an assay for “total” hepatitis C core antigen. J Virol Methods. 2001;96(2):127–32.
Non-commercial or off-market assay [
PubMed: 11445143]
- 94.
Lagging
LM
, GarciaCE, WestinJ, WejstålR, NorkransG, DhillonAP, et al. Comparison of serum hepatitis C virus RNA and core antigen concentrations and determination of whether levels are associated with liver histology or affected by specimen storage time. J Clin Microbiol. 2002;40(11):4224–9.
Non-commercial or off-market assay [
PMC free article: PMC139660] [
PubMed: 12409402]
- 95.
Lambert
N
. Value of HCV antigen-antibody combined HCV assay in hepatitis C diagnosis. Dev Biol (Basel). 2007;127:113–21.
Review article [
PubMed: 17486884]
- 96.
Laperche
S
, ElghouzziMH, MorelP, Asso-BonnetM, Le MarrecN, GiraultA, et al. Is an assay for simultaneous detection of hepatitis C virus core antigen and antibody a valuable alternative to nucleic acid testing?Transfusion. 2005;45(12):1965–72.
Non-human subjects or commercial samples [
PubMed: 16371051]
- 97.
Combined detection of hepatitis C virus core antigen and antibody as an alternative to nucleic acid testing in blood screening. Vox Sang. 2005;89:21. Abstract or poster
- 98.
Laperche
S
, Le MarrecN, SimonN, BouchardeauF, DeferC, Maniez-MontreuilM, et al. A new HCV core antigen assay based on disassociation of immune complexes: an alternative to molecular biology in the diagnosis of early HCV infection. Transfusion. 2003;43(7):958–62.
Noncommercial [
PubMed: 12823757]
- 99.
Laperche
S
, RougerP, SmiloviciW, HerveP, LefrereJJ. Alternatives to nucleic acid testing in the blood transfusion service. Lancet. 2002;360(9344):1519.
Editorial or comment [
PubMed: 12433563]
- 100.
Larrat
S
, BourdonC, BaccardM, GarnaudC, HilleretMN, QuesadaJL, et al. HCV screening without venipuncture: pointof-care versus laboratory-based antigenantibody combined assay. Hepatology. 2011;54:573A–4A. Abstract or poster
- 101.
Larrat
S
, BourdonC, BaccardM, GarnaudC, MathieuS, QuesadaJL, et al. Performance of an antigen-antibody combined assay for hepatitis C virus testing without venipuncture. J Clin Virol. 2012;55(3):220–5.
Inappropriate reference test [
PubMed: 22901327]
- 102.
Leary
TP
, GutierrezRA, MuerhoffAS, BirkenmeyerLG, DesaiSM, DawsonGJ. A chemiluminescent, magnetic particle-based immunoassay for the detection of hepatitis C virus core antigen in human serum or plasma. J Med Virol. 2006;78(11):1436–40.
Non-human subjects or commercial samples [
PubMed: 16998880]
- 103.
Lee
L
, PerrettG, HowardP, MakanjuolaD, ClarkJ. HCV p22 antigen test: Serological response and diagnostic advantages. Gut. 2012;61:A140. Abstract or poster
- 104.
Lee
S
, KimYS, JoMJ, JinM, LeeDK, KimS. Chip-based detection of hepatitis C virus using RNA aptamers that specifically bind to HCV core antigen. Biochem Biophys Res Commun. 2007;358(1):47–52.
Non-commercial or off-market assay [
PubMed: 17475212]
- 105.
Lee
SR
, PetersonJ, NivenP, BahlC, PageE, DeLeysR, et al. Efficacy of a hepatitis C virus core antigen enzyme-linked immunosorbent assay for the identification of “window-phase” blood donations. Vox Sang. 2001;80(1):19–23.
Non-commercial or off-market assay [
PubMed: 11339063]
- 106.
Leon
P
, LopezJA, ElolaC, LeeSR, CalmannM, EchevarriaJM. Use of overlapping synthetic peptides to characterize samples from blood donors with indeterminate results to hepatitis C virus core antigen. Vox Sang. 1998;75(1):32–6.
No HCV core antigen performed [
PubMed: 9745151]
- 107.
Letowska
M
, RosiekA, GronowskaA, MikulskaM, GrabarczykP, BrojerE. Hepatitis C virus (HCV) core antigen detection in HCV RNA-positive/anti-HCV-negative Polish blood donors identified by nucleic acid testing. Transfusion. 2009;49(10):2241–2.
Editorial or comment [
PubMed: 19903286]
- 108.
Li Cavoli
G
, ZagarrigoC, SchillaciO, TralongoA, RotoloU. Hepatitis C virus core antigen testing in the monitoring of patients on dialysis. Saudi J Kidney Dis Transpl. 2012;23(5):1056–8.
Editorial or comment [
PubMed: 22982924]
- 109.
Li Cavoli
G
, ZagarrigoC, TralongoA, SchillaciO, RotoloU, Li DestriN, et al. Hepatitis C virus core antigen in virological monitoring of dialysis patients. NDT Plus. 2010;3:iii247.
Duplicate data [
PMC free article: PMC5477968] [
PubMed: 28657071]
- 110.
Liao
WS
, ShenCH, TungSY, LeeJH. Detection of NS3 antigen of hepatitis C virus in liver tissue and its relation to serum HCV-RNA. J Gastroenterol Hepatol. 2009;24:A182. Non-blood specimen
- 111.
Long
L
, ShenT, GaoJ, DuanZ, LiangH, LuF. Effectiveness of HCV core antigen and RNA quantification in HCV-infected and HCV/HIV-1-coinfected patients. BMC Infect Dis. 2014;14:577.
No extractable data, non-response from author [
PMC free article: PMC4225041] [
PubMed: 25371245]
- 112.
Long
RX
, LiH, CuiPF. Stability of three hepatitis C virus markers. Chinese Journal of Biologicals. 2009;22(5):468–9, 473. No HCV core antigen performed
- 113.
Lorenzo
J
, CastroA, AguileraA, PrietoE, López-CalvoS, RegueiroB, et al. Total HCV core antigen assay: a new marker of HCV viremia and its application during treatment of chronic hepatitis C. J Virol Methods. 2004;120(2):173–7.
Non-commercial or off-market assay [
PubMed: 15288960]
- 114.
Lozano
ML
, CandelaMJ, LozanoML, CanoH, ZuazuI, VicenteV. Detection of free hepatitis C virus core antigen by enzyme-linked immunosorbent assay is not suitable for screening of granulocyte colony-stimulating factor-mobilized hematopoietic progenitor donors. Transfusion. 2004;44(12):1755–61.
Non-commercial or off-market assay [
PubMed: 15584991]
- 115.
Łucejko
M
, GrzeszczukA, JaroszewiczJ, FlisiakR. [Serum HCV core antigen concentration in HCV monoinfection and HCV/HIV coinfection.] Pol Merkur Lekarski. 2013;35(206):72–6.
Unable to translate [
PubMed: 24052984]
- 116.
Lunel
F
, PivertA, PayanC, CommRS. Comparison of HCV RNA and HCV core antigen kinetics in the follow up of a therapeutic protocol in HCV-HIV co-infected patients (RIBAVIC). J Hepatol. 2004;40:144–5. Abstract or poster
- 117.
Lunel
F
, VeillonP. Antigen and viral load. Transfus Clin Biol. 2003;10(2):74–7.
Non-commercial or off-market assay [
PubMed: 12763148]
- 118.
Lunel
F
, VeillonP, PayanC. Evaluation of the ortho total HCV core antigen assay in comparison to methods of detection and quantification for HCV RNA. Hepatology. 2002;36(4):353A. Abstract or poster
- 119.
Lunel-Fabiani
F
, PayanC. Virological tools for the diagnosis and follow up of hepatitis C: use and role of new tests. Gastroenterol Clin Biol. 2003;27(8–9):718–26.
Review article [
PubMed: 14586244]
- 120.
Ma
CX
, XieGM, ZhangW, LiangM, LiuB, XiangH. Label-free sandwich type of immunosensor for hepatitis C virus core antigen based on the use of gold nanoparticles on a nanostructured metal oxide surface. Mikrochim Acta. 2012;178(3–4):331–40. Non-commercial or off-market assay
- 121.
Masalova
OV
, AtanadzeSN, SamokhvalovEI, PetrakovaNV, KalininaTI, SmirnovVD, et al. Detection of hepatitis C virus core protein circulating within different virus particle populations. J Med Virol. 1998;55(1):1–6.
Non-commercial or off-market assay [
PubMed: 9580878]
- 122.
Masalova
OV
, VishnevskaiaTV, ShkurkoTV, GaranzhaTA, TupolevaTA, FilatovFP, et al. Comparative analysis of hepatitis C virus core protein in the plasma and serum samples from HCV-infected blood donors and patients with hepatitis C. Vopr Virusol. 2007;52(4):11–7.
Noncommercial or off-market assay [
PubMed: 17722604]
- 123.
Masopust
J
, KracíkováJ, NěmečekV, ProcházkováR. [A combined antigen-antibody HCV detection assay.] Transfuze a Hematologie Dnes. 2005;11(4):141–7. Unable to translate
- 124.
Massaguer
A
, FornsX, CostaJ, FeliuA, García-RetortilloM, NavasaM, et al. Performance of hepatitis C virus core antigen immunoassay in monitoring viral load after liver transplantation. Transplantation. 2005;79(10):1441–4.
Non-commercial or off-market assay [
PubMed: 15912117]
- 125.
Mathur
A
. Comparative evaluation of anti-HCV-third-generation assay and anti-HCV-fourth-generation assay in blood donor screening. Transfusion. 2014;54:216A–7A. Abstract or poster
- 126.
Mayerhofer
T
, DeimelM, AmbrusE, BertschAL, AliskanovicV, Trubert-ExingerD, et al. Evaluation of the clinical performance of the Abbott ARCHITECT HCV core antigen assay. Clinical Chemistry and Laboratory Medicine. 2012;50(4):A86. Abstract or poster
- 127.
Mederacke
I
, MeierM, LüthJB, Schmidt-GürtlerH, RaupachR, Horn-WichmannR, et al. Different kinetics of HBV and HCV during haemodialysis and absence of seronegative viral hepatitis in patients with end-stage renal disease. Nephrol Dial Transplant. 2011;26(8):2648–56.
Less than 10 independent samples [
PubMed: 21273235]
- 128.
Mederacke
I
, RaupachR, MannsMP, WedemeyerH, TillmannHL. Quantitative HCV core antigen and HCV-RNA kinetics in HCV infected patients over a period of up to 8 years. Hepatology. 2009;50(4):1070A–1A. Abstract or poster
- 129.
Medhi
S
, PotukuchiSK, PolipalliSK, SwargiarySS, DekaP, ChaudharyA, et al. Diagnostic utility of hepatitis C virus core antigen in hemodialysis patients. Clin Biochem. 2008;41(7–8):447–52.
Non-commercial or off market assay [
PubMed: 18267117]
- 130.
Meng
S
, LiX, YinH, LiDF. Establishment of detection test for hepatitis C virus antigen. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2001;15(3):287–90.
Non-commercial or off-market assay [
PubMed: 11986709]
- 131.
Miceli
M
, AgrestiA, PalangeM, IudiconeP, PaluzziC, PerretM, et al. A new high sensitive HCV antigen assay for blood donors screening. Vox Sang. 2010;99:300. Abstract or poster
- 132.
Mihaljevic
I
. Evaluation of the architect HCV antigen assay for use in organ and tissue donors. Vox Sang. 2009;96:24. Abstract or poster
- 133.
Moini
M
, ZiyaeyanM, AghaeiS, SaghebMM, TaghaviSA, MoeiniM, et al. Hepatitis C virus (HCV) infection rate among seronegative hemodialysis patients screened by two methods; HCV core antigen and polymerase chain reaction. Hepat Mon. 2013;13(6).
Non-commercial or off-market assay [
PMC free article: PMC3768235] [
PubMed: 24032048]
- 134.
Moini
M
, ZiyaeyanM, AghaeiS, SaghebMM, TaghaviSA, MoeiniM, et al. Hepatitis C virus (HCV) screening using HCV core antigen in hemodialysis patients. Hepatol Int. 2012;6(1):150.
Abstract or poster [
PMC free article: PMC3768235] [
PubMed: 24032048]
- 135.
Moreno
M
, Pérez-AlvarezR, RodrigoL, Pérez-LópezR, Suárez-LeivaP. Long-term evolution of serum and liver viral markers in patients treated for chronic hepatitis C and sustained response. J Viral Hepat. 2006;13(1):28–33.
Non-commercial or off market-assay [
PubMed: 16364079]
- 136.
Moriya
T
, SasakiF, TanakaJ, MizuiMa, NakanishiT, TakahashiK, et al. Comparison of HCV core antigen activity by ELISA and amount of HCV RNA by branched DNA assay. International Hepatology Communications. 1994;2(3):175–7. Non-commercial or off-market assay
- 137.
Morota
K
, FujinamiR, KinukawaH, MachidaT, OhnoK, SaegusaH, et al. A new sensitive and automated chemiluminescent microparticle immunoassay for quantitative determination of hepatitis C virus core antigen. J Virol Methods. 2009;157(1):8–14.
Non-human subjects or commercial samples [
PubMed: 19135481]
- 138.
Muerhoff
AS
, JiangL, ShahDO, GutierrezRA, PatelJ, GarolisC, et al. Detection of HCV core antigen in human serum and plasma with an automated chemiluminescent immunoassay. Transfusion. 2002;42(3):349–56.
Non-commercial or off-market assay [
PubMed: 11961241]
- 139.
Mukomolov
S
, BarantsevichE, MukomolovaA, BarantsevichN, SchlyakhtoE. Study of sensitivity and specificity of ELISA test system MONOLISA HCV Ag-Ab ULTRA. Clin Microbiol Infect. 2010;16:S298. Abstract or poster
- 140.
Nayak
NC
, SatharSA. Immunohistochemical detection of hepatitis C virus antigen in paraffin embedded liver biopsies from patients with chronic liver disease. Acta Histochem. 1999;101(4):409–19.
Non-blood specimens [
PubMed: 10611929]
- 141.
Netski
DM
, WangXH, MehtaSH, NelsonK, CelentanoD, ThongsawatS, et al. Hepatitis C virus (HCV) core antigen assay to detect ongoing HCV infection in Thai injection drug users. J Clin Microbiol. 2004;42(4):1631–6.
Non-commercial or off-market assay [
PMC free article: PMC387596] [
PubMed: 15071017]
- 142.
Nuovo
GJ
, HollyA, WakelyP, FrankelW. Correlation of histology, viral load, and in situ viral detection in hepatic biopsies from patients with liver transplants secondary to hepatitis C infection. Hum Pathol. 2002;33(3):277–84.
Non-blood specimens [
PubMed: 11979367]
- 143.
Oliva
JA
, ErcillaG, MallafreJM, BrugueraM, CarrioJ, PereiraBJ. Markers of hepatitis C infection among hemodialysis patients with acute and chronic infection: implications for infection control strategies in hemodialysis units. Int J Artif Organs. 1995;18(2):73–7.
No HCV core antigen performed [
PubMed: 7558399]
- 144.
Orito
E
, MizokamiM, TanakaT, LauJYN, SuzukiK, YamauchiM, et al. Quantification of serum hepatitis C virus core protein level in patients chronically infected with different hepatitis C virus genotypes. Gut. 1996;39(6):876–80.
Non-commercial or off-market assay [
PMC free article: PMC1383464] [
PubMed: 9038674]
- 145.
Ottiger
C
, GygliN, HuberAR. Detection limit of architect hepatitis C Core antigen assay in correlation with HCV-PCR viral load. Clinical Chemistry and Laboratory Medicine. 2012;50(5):A171. Abstract or poster
- 146.
Pawelczyk
A
, KubisaN, JablonskaJ, Bukowska-OskoI, CaraballoCortes K, FicM, et al. Detection of hepatitis C virus (HCV) negative strand RNA and NS3 protein in peripheral blood mononuclear cells (PBMC): CD3+, CD14+ and CD19+. Virol J. 2013;10:346.
No HCV core antigen performed [
PMC free article: PMC4222874] [
PubMed: 24279719]
- 147.
Pawlotsky
JM
. Use and interpretation of virological tests for hepatitis C. Hepatology. 2002;36(5):S65–S73.
Review article [
PubMed: 12407578]
- 148.
Pawlotsky
JM
. Use and interpretation of hepatitis C virus diagnostic assays. Clin Liver Dis. 2003;7(1):127–37.
Review article [
PubMed: 12691462]
- 149.
Pessôa
MG
, AlvesVAF, WakamatsuA, GomesJG, MaertensG, van der BorghtB, et al. Post-transplant recurrent hepatitis C: immunohistochemical detection of hepatitis C virus core antigen and possible pathogenic implications. Liver Int. 2008;28(6):807–13.
Non-blood specimens [
PubMed: 18422936]
- 150.
Peterson
J
, GreenG, IidaK, CaldwellB, KerrisonP, BernichS, et al. Detection of hepatitis C core antigen in the antibody negative “window” phase of hepatitis C infection. Vox Sang. 2000;78(2):80–5.
Non-commercial or off-market assay [
PubMed: 10765142]
- 151.
Pham
BN
, Martinot-PeignouxM, RipaultMP, BoyerN, LevyV, MarcellinP. Quantitative measurement of hepatitis C virus core antigen is affected by the presence of cryoglobulins. Clin Exp Immunol. 2006;146(2):211–17.
Non-commercial or off-market assay [
PMC free article: PMC1942051] [
PubMed: 17034572]
- 152.
Pivert
A
, PayanC, LunelF, Conseil scientifique R. Comparison of hepatitis C viral RNA and core antigen kinetics in the therapeutic follow up of hepatitis C virus and human immunodeficiency virus co-infected patients, treated by bitherapy interferon-ribavirin, within the framework of RIBAVIC protocol. Pathol Biol (Paris). 2004;52(9):522–8.
Duplicate data [
PubMed: 15531116]
- 153.
Podesta
MA
, CancariniG, CucchiariD, MontanelliA, BadalamentiS, GrazianiG. Diagnosis and follow-up of HCV infection in hemodialysis patients and renal transplant recipients: HCV core antigen and IgM anti-HCV. Nephrol Dial Transplant. 2014;29:iii514. Abstract or poster
- 154.
Poljak
M
, LepejSZ, RodeOA. Recent developments in serologic and molecular diagnosis of hepatitis B and C. Acta Med Croatica. 2013;67(4):281–90.
Review article [
PubMed: 24984327]
- 155.
Pontisso
P
, CarabaichA, BernardinelloE, BoccatoS, ChemelloL, GattaA, et al. Correlation of HCV RNA to total HCV core antigen in chronic hepatitis C patients. Hepatology. 2002;36(4):546A. Abstract or poster
- 156.
Pradat
P
, MaynardM, BerthillonP, BaillyF, BordesI, TillmannHL, et al. Baseline HCV core antigen/HCV RNA ratio: association with treatment responses. Hepatology. 2004;40(4):326A. Abstract or poster
- 157.
Raffaele
L
, SpreaficoM, FoglieniB, GuarnoriI, AlessandraB, GalliC, et al. Performance evaluation of fully automated quantitative HCV-core antigen assay. Transfusion. 2010;50:202A. Abstract or poster
- 158.
Raker
CA
, TaborE, OkayamaA, YuMYW, KoharaM, MuellerNE, et al. HCV core antigen as an alternative to NAT to detect HCV viremia. Transfusion. 2004;44(2):307–8.
Editorial or comment [
PubMed: 14962325]
- 159.
Ravera
G
, BottaroLC, FranceschiniM, MorandoA, De PoloM, ZareM, et al. Reliability and diagnostic use of a test for the search of the hepatitis C virus Ag (AgHCV). Hepatogastroenterology. 2006;53(71):753–6.
Non-commercial or off-market assay [
PubMed: 17086882]
- 160.
Rebucci
C
, CerinoA, CividiniA, TimoL, FurioneM, MondelliMU. Monitoring response to antiviral therapy for patients with chronic hepatitis C virus infection by a core-antigen assay. J Clin Microbiol. 2003;41(8):3881–4.
Less than 10 independent samples [
PMC free article: PMC179816] [
PubMed: 12904409]
- 161.
Reddy
AK
, DakshinamurtyKV, LakshmiV. Utility of HCV core antigen ELISA in the screening for hepatitis C virus infection in patients on hemodialysis. Indian J Med Microbiol. 2006;24(1):55–7.
Less than 10 independent samples [
PubMed: 16505558]
- 162.
Rehany
U
, ChaikovskyI, Kra-OzZ, SatingerJ, BersudskyV, CohenI. The expression of hepatitis-C virus antigen by immunohistochemical stain and polymerase chain reaction in corneas of seropositive corneal donors. Cell Tissue Bank. 2002;3(2):139–44.
Non-blood specimens [
PubMed: 15256891]
- 163.
Romano
L
, ZanettiAR, BrunettoM, CiccorossiP, LeeSR, CalmannM, et al. Correlation of total HCV core antigen ELISA to HCV RNA in liver transplant patients. Hepatology. 2002;36(4):654A. Abstract or poster
- 164.
Ross
RS
, ViazovS, SalloumS, HilgardP, GerkenG, RoggendorfM. Analytical performance characteristics and clinical utility of a novel assay for total hepatitis C virus core antigen quantification. J Clin Microbiol. 2010;48(4):1161–8.
No extractable data, no response from author [
PMC free article: PMC2849592] [
PubMed: 20107102]
- 165.
Sabry
AA
, SobhMA, IrvingWL, GrabowskaA, WagnerBE, FoxS, et al. A comprehensive study of the association between hepatitis C virus and glomerulopathy. Nephrol Dial Transplant. 2002;17(2):239–45.
Non-blood specimens [
PubMed: 11812873]
- 166.
Saeed
M
, SuzukiR, KondoM, AizakiH, KatoT, MizuochiT, et al. Evaluation of hepatitis C virus core antigen assays in detecting recombinant viral antigens of various genotypes. J Clin Microbiol. 2009;47(12):4141–3.
Non-human subjects or commercial samples [
PMC free article: PMC2786616] [
PubMed: 19812276]
- 167.
Sansonno
D
, IacobelliAR, CornacchiuloV, IodiceG, DammaccoF. Detection of hepatitis C virus (HCV) proteins by immunofluorescence and HCV RNA genomic sequences by non isotopic in situ hybridization in bone marrow and peripheral blood mononuclear cells of chronically HCV-infected patients. Clin Exp Immunol. 1996;103(3):414–21.
Non-blood specimens [
PMC free article: PMC2200370] [
PubMed: 8608640]
- 168.
Sansonno
D
, LaulettaG, DammaccoF. Detection and quantitation of HCV core protein in single hepatocytes by means of laser capture microdissection and enzyme-linked immunosorbent assay. J Viral Hepat. 2004;11(1):27–32.
Non-blood specimens [
PubMed: 14738555]
- 169.
Sansonno
D
, LaulettaG, MontroneM, GrandalianoG, SchenaFP, DammaccoF. Hepatitis C virus RNA and core protein in kidney glomerular and tubular structures isolated with laser capture microdissection. Clin Exp Immunol. 2005;140(3):498–506.
Non-blood specimens [
PMC free article: PMC1809381] [
PubMed: 15932511]
- 170.
Sansonno
D
, LaulettaG, NisiL, GattiP, PesolaF, PansiniN, et al. Non-enveloped HCV core protein as constitutive antigen of cold-precipitable immune complexes in type II mixed cryoglobulinaemia. Clin Exp Immunol. 2003;133(2):275–82.
Non-commercial or off-market assay [
PMC free article: PMC1808767] [
PubMed: 12869035]
- 171.
Sansonno
L
, LaulettaG, RussiS, DammaccoF. In situ simultaneous detection of hepatitis C virus RNA and hepatitis C virus-related antigens in hepatocellular carcinoma. Cancer Treatment Reviews. 2010;36:S98.
Abstract or poster [
PubMed: 9210705]
- 172.
Sarov
B
, NovackL, BeerN, SafiJ, SolimanH, PliskinJS, et al. Feasibility and cost-benefit of implementing pooled screening for HCVAg in small blood bank settings. Transfus Med. 2007;17(6):479–87.
Does not apply to study question [
PubMed: 17727618]
- 173.
Sarrazin
C
. Advances in diagnosis and management of virus c hepatitis. Biochim Clin. 2013;37:S60–S1. Review article
- 174.
Schiano
TD
, GutierrezJA, WalewskiJL, FielMI, ChengB, BodenheimerJrH, et al. Accelerated hepatitis C virus kinetics but similar survival rates in recipients of liver grafts from living versus deceased donors. Hepatology. 2005;42(6):1420–8.
Does not apply to study question, noncommercial or off-market assay [
PubMed: 16317672]
- 175.
Schiano
TD
, GutierrezJA, WalewskiJL, FielMI, ChengB, NelsonC, et al. HCV RNA, core antigen, ALT values, and histology after liver transplantation: two year follow-up of CDLT versus LDLT. Hepatology. 2004;40(4):164A. Abstract or poster
- 176.
Schmidt
AJ
, FalcatoL, ZahnoB, BurriA, RegenassS, MüllhauptB, et al. Prevalence of hepatitis C in a Swiss sample of men who have sex with men: Whom to screen for HCV infection? BMC Public Health. 2014;14:3.
Less than 10 independent samples [
PMC free article: PMC3890510] [
PubMed: 24393532]
- 177.
Schüttler
CG
, ThomasC, DischerT, FrieseG, LohmeyerJ, SchusterR, et al. Variable ratio of hepatitis C virus RNA to viral core antigen in patient sera. J Clin Microbiol. 2004;42(5):1977–81.
Non-commercial or off-market assay [
PMC free article: PMC404599] [
PubMed: 15131157]
- 178.
Seiskari
T
, HorstiJ, AittoniemiJ. The value of HCV antigen test with reference to PCR in confirming infection in cases with indefinite antibody test result. J Clin Virol. 2011;51(1):90–1.
Editorial or comment [
PubMed: 21377410]
- 179.
Seme
K
, PoljakM, BabicDZ, MocilnikT, VinceA. The role of core antigen detection in management of hepatitis C: a critical review. J Clin Virol. 2005;32(2):92–101.
Review article [
PubMed: 15653411]
- 180.
Sentjens
RE
, van BaalK, VerhoevenG, LeeS, PieksmaF, NivenP, et al. Comparison of first and second generation HCV core antigen test with qualitative and quantitative HCV-RNA PCR. Hepatology. 2000;32(4):548A. Abstract or poster
- 181.
Serdarevic
N
. Detection of hepatitis C in serum using anti HCV and HCV Ag. Clinical Chemistry and Laboratory Medicine. 2014;52:S975. Abstract or poster
- 182.
Shah
DO
, ChangC, GutierrezR, ChengKY, JiangLX, SalbillaV, et al. Hepatitis C virus antibody test on a chemiluminescence automated analyzer as an aid in the diagnosis of HCV infection and for blood screening. Transfusion. 2010;50:203A–4A. Abstract or poster
- 183.
Shah
DO
, ChangCD, JiangLX, ChengKY, MuerhoffAS, GutierrezRA, et al. Combination HCV core antigen and antibody assay on a fully automated chemiluminescence analyzer. Transfusion. 2003;43(8):1067–74.
Non-commercial or off-market assay [
PubMed: 12869112]
- 184.
Shah
O
, ChangD, GutierrezA, ChengY, DesaiM, DawsonJ. Hepatitis C virus antibody assay on a chemiluminescence automated analyzer as an AID in the diagnosis of HCV infection and for blood screening. Vox Sang. 2010;99:297–8. Abstract or poster
- 185.
Shaw
SG
, CresswellF, HomerG, Hassan-IbrahimM, FisherM. Hepatitis C antigen testing: a reliable alternative for diagnosing acute HCV infection. Top Antivir Med. 2014;22(e–1):312. Abstract or poster
- 186.
Shen
T
, ChenX, ZhangW, XiY, CaoG, ZhiY, et al. A higher correlation of HCV core antigen with CD4+ T cell counts compared with HCV RNA in HCV/HIV coinfected patients. Hepatol Int. 2012;6(1):150.
Abstract or poster [
PMC free article: PMC3155566] [
PubMed: 21858166]
- 187.
Shepherd
SJ
, AitkenC, WalkowiczM, McOwanJ, CameronSO, CarmanWF. HCV antigen testing in a busy diagnostic laboratory. Clin Microbiol Infect. 2012;18:676–7. Abstract or poster
- 188.
Shi
HB
, XieL, HuangDZ, HeLX. Detection of HCV antigen in the serum by immunodotting. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2006;22(3):336–8.
Non-commercial or off market assay [
PubMed: 16643794]
- 189.
Song
D
, KangJE, KimSY, HwangSH, KimHH, LeeEY, et al. [Evaluation of ARCHITECT HCV core antigen assay.] [Article in Korean] Korean J Lab Med. 2010;30(6):654–9.
Unable to translate [
PubMed: 21157153]
- 190.
Syed
SI
, SadiqS. Immunohistochemical detection of hepatitis C virus (HCV) in liver biopsies of hepatitis C patients. J Pak Med Assoc. 2011;61(12):1198–201.
Non-blood specimens [
PubMed: 22355966]
- 191.
Tagny
CT
, MbanyaD, MurphyEL, LefrèreJJ, LapercheS. Screening for hepatitis C virus infection in a high prevalence country by an antigen/antibody combination assay versus a rapid test. J Virol Methods. 2014;199:119–23.
Inappropriate reference test [
PMC free article: PMC5042449] [
PubMed: 24487098]
- 192.
Takahashi
K
, OkamotoH, KishimotoS, MunekataE, TachibanaK, AkahaneY, et al. Demonstration of a hepatitis C virus-specific antigen predicted from the putative core gene in the circulation of infected hosts. J Gen Virol. 1992;73(Pt 3):667–72.
No HCV core antigen performed [
PubMed: 1312124]
- 193.
Tanaka
E
, KiyosawaK, MatsumotoA, KashiwakumaT, HasegawaA, MoriH, et al. Serum levels of hepatitis C virus core protein in patients with chronic hepatitis C treated with interferon alfa. Hepatology. 1996;23(6):1330–3.
Non-commercial or off-market assay [
PubMed: 8675147]
- 194.
Tanaka
E
, OhueC, AoyagiK, YamaguchiK, YagiS, KiyosawaK, et al. Evaluation of a new enzyme immunoassay for hepatitis C virus (HCV) core antigen with clinical sensitivity approximating that of genomic amplification of HCV RNA. Hepatology. 2000;32(2):388–93.
Non-commercial or off market assay [
PubMed: 10915747]
- 195.
Tanoue
Y
, MaekawaH, InoueT, WatanabeT, ShimodaH, KurodaT, et al. The Cobas AmpliPrep/Cobas TaqMan real-time polymerase chain reaction assay produced false-negative results in two patients with hepatitis C virus genotype 2. Kanzo. 2013;54(7):507–8. Less than 10 independent samples
- 196.
Teo
CG
. Changing strategies for hepatitis C testing. Antivir Ther. 2012;17(7):1391–5.
Review article [
PubMed: 23322655]
- 197.
Thong
V
, AkkarathamrongsinS, AvihingsanonA, PoovorawanY, TangkijvanichP. The correlation between hepatitis C core antigen and HCV RNA levels with respect to HIV status, HCV genotype and IFNL4 polymorphism. J Viral Hepat. 2014;21:21. Abstract or poster
- 198.
- 199.
Tillmann
HL
, WiegandJ, GlombI, JelineckA, PicchioG, WedemeyerH, et al. Diagnostic algorithm for chronic hepatitis C virus infection: role of the new HCV-Core antigen assay. Z Gastroenterol. 2005;43(1):11–6.
Non-commercial or off-market assay [
PubMed: 15650966]
- 200.
Tobler
LH
, StramerSL, LeeSR, BaggettD, WrightD, HirschkornD, et al. Performance of ORTHO (R) HCV core antigen and trak-C (TM) assays for detection of viraemia in pre-seroconversion plasma and whole blood donors. Vox Sang. 2005;89(4):201–7.
Non-commercial or off-market assay [
PubMed: 16262752]
- 201.
Tokita
H
, KaufmannGR, MatsubayashiM, OkudaI, TanakaT, HardaH, et al. Hepatitis C virus core mutations reduce the sensitivity of a fluorescence enzyme immunoassay. J Clin Microbiol. 2000;38(9):3450–2.
Non-commercial or off-market assay [
PMC free article: PMC87404] [
PubMed: 10970401]
- 202.
Tsutsumi
M
, UrashimaS, TakadaA, DateT, TanakaY. Detection of antigens related to hepatitis C virus RNA encoding the NS5 region in the livers of patients with chronic type C hepatitis. Hepatology. 1994;19(2):265–72.
Non-blood specimens [
PubMed: 7507461]
- 203.
Valcavi
P
, MediciMC, CasulaF, ArcangelettiMC, De ContoF, PinardiF, et al. Evaluation of a total hepatitis C virus (HCV) core antigen assay for the detection of antigenaemia in anti-HCV positive individuals. J Med Virol. 2004;73(3):397–403.
Non-commercial or off-market assay [
PubMed: 15170635]
- 204.
Vanhommerig
JW
, Van De LaarTJW, Van RooijenMS, De VriesHJ, SpeksnijderAGCL, PrinsM, et al. Hepatitis c virus (HCV) core antigen assay for detection of active HCV infection among HIV-infected men who have sex with men attending an STI clinic. J Hepatol. 2014;60(1):S312. Duplicate data
- 205.
Vargas
V
, KrawczynskiK, CastellsL, MartinezN, EstebanJ, AllendeH, et al. Recurrent hepatitis C virus infection after liver transplantation: Immunohistochemical assessment of the viral antigen. Liver Transpl Surg. 1998;4(4):320–7.
Non-blood specimens [
PubMed: 9649647]
- 206.
Veillon
P
, PayanC, PicchioG, Maniez-MontreuilM, GuntzP, LunelF. Comparative evaluation of the total hepatitis C virus core antigen, branched-DNA, and amplicor monitor assays in determining viremia for patients with chronic hepatitis C during interferon plus ribavirin combination therapy. J Clin Microbiol. 2003;41(7):3212–20.
Non-commercial or off-market assay [
PMC free article: PMC165326] [
PubMed: 12843066]
- 207.
Vrielink
H
, VanderpoelCL, ReesinkHW, LeliePN. Comparison of two anti-hepatitis C virus enzyme-linked immunosorbent assays: Wellcozyme-VK45 and Ortho-2.0. Infusionsther Transfusionsmed. 1995;22(3):164–7.
No HCV core antigen performed [
PubMed: 7543783]
- 208.
Vucetic
D
, TrkuljicM, BalintB, BorovcaninN, LjubenovM, JovicicD. Evaluation of immunoassays for hepatitis C Virus antigen and antibody combined detection in Reactive serum samples among blood donors. Vox Sang. 2009;96:95. Abstract or poster
- 209.
Waldenström
J
, KonarJ, EkermoB, NorderH, LaggingM. Neonatal transfusion-transmitted hepatitis C virus infection following a pre-seroconversion window-phase donation in Sweden. Scand J Infect Dis. 2013;45(10):796–9.
Less than 10 independent samples [
PubMed: 23746339]
- 210.
Wang
C
, ZhangL, ShenX. Development of a nucleic acid lateral flow strip for detection of hepatitis C virus (HCV) core antigen. Nucleosides Nucleotides Nucleic Acids. 2013;32(2):59–68.
Non-commercial or off-market assay [
PubMed: 23448141]
- 211.
Wang
F
, WangS, JinL. Detections of hepatitis C virus RNA and NS3 antigen and their relation to liver histopathology. Zhonghua Yi Xue Za Zhi. 1995;75(11):670–2, 709–10.
Non-blood specimens [
PubMed: 8697087]
- 212.
Wang
FC
, ShiZY, CaiJ, SuJ. Evaluation on the use of detection of hepatitis C core antigen for screening blood donor. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2007;21(4):389–90.
Less than 10 independent samples [
PubMed: 18322613]
- 213.
Watanabe
J
, MatsumotoC, FujimuraK, ShimadaT, YoshizawaH, OkamotoH, et al. Predictive value of screening tests for persistent hepatitis C virus infection evidenced by viraemia. Japanese experience. Vox Sang. 1993;65(3):199–203.
No HCV core antigen performed [
PubMed: 7504373]
- 214.
Widell
A
, MolnegrenV, PieksmaF, CalmannM, PetersonJ, LeeSR. Detection of hepatitis C core antigen in serum or plasma as a marker of hepatitis C viraemia in the serological window-phase. Transfus Med. 2002;12(2):107–13.
Non-commercial or off-market assay [
PubMed: 11982963]
- 215.
Willems
M
, LerutJ, RoskamsT, DonataccioM, CiccarelliO, HabetsW, et al. Detection of HCV antigen and HCV-RNA in liver biopsies of patients with chronic hepatitis-C undergoing liver-transplantation – a close correlation. Hepatology. 1993;18(4):A262–A. Non-blood specimens
- 216.
Wong
JSJ
, KaramalakisD. Evaluation of automated hepatitis C core antigen assay in occupational exposure. Pathology. 2013;45(5):529–31.
Editorial or comment [
PubMed: 23856850]
- 217.
Wu
RH
. [Detection of hepatitis C virus antigen in hemodialysis patients]. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2009;23(3):232–4.
Review article [
PubMed: 20104788]
- 218.
Xie
L
, GuangYP, LiuF, ShiHB, YanL, LouJL. A magnetic microparticle-based immunoassay for hepatitis C virus NS3 antigen. Afr J Microbiol Res. 2011;5(1):28–33. Non-commercial or off-market assay
- 219.
Xie
L
, HuangDZ, ChenHL, HeLX, WangJ, HanDK. The clinical application and analysis of hepatitis C virus NS 3 antigen detection by ELISA in human serum. Chinese Journal of Microbiology and Immunology. 2009;29(1):88–92.
No extractable data, non-response from author [
PubMed: 17374280]
- 220.
Xie
L
, WuXD, HuangDZ, ChenHL, HeLX, WangJ, et al. Clinical application and analysis of hepatitis C virus NS3 antigen detection by ELISA in human serum. Chin Med J (Engl).
Duplciate data2007;120(4):294–9. [
PubMed: 17374280]
- 221.
Yagci
S
, PadalkoE. Comparison of monolisa HCV Ag/Ab ULTRA with two anti-HCV assays for the detection of HCV infection in hospital setting. Curr Microbiol. 2012;64(2):148–51.
Inappropriate reference test [
PubMed: 22076114]
- 222.
Yan
F
, HaoF, ZhaoL. Study of expression of hepatitis C virus antigens and viral replication in extrahepatic tissues. Zhonghua Gan Zang Bing Za Zhi. 2000;8(1):40–2.
Non-blood specimens [
PubMed: 10712785]
- 223.
Yan
FM
, ChenAS, HaoF, ZhaoXP, GuCH, ZhaoLB, et al. Hepatitis C virus may infect extrahepatic tissues in patients with hepatitis C. World J Gastroenterol. 2000;6(6):805–11.
Non-blood specimens [
PMC free article: PMC4728266] [
PubMed: 11819700]
- 224.
Yang
RF
, WeiL. Identifying factors that may influence utility of HCV core antigen assay. Hepatol Int. 2014;8(1):S200. Abstract or poster
- 225.
Yang
Z
, QiZB, YuY. Evaluation of A kit for determination of total hepatitis C virus core antigen. Chinese Journal of Biologicals. 2008;21(1):51–3. Non-commercial or off-market assay
- 226.
Yao
RN
, ZhangJH, HuangXJ, YangQ, CaoQ, JiangXC. Technique of detection of hepatitis C core antigen used in safety blood transfusion. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2006;14(3):617–8.
Non-commercial or off-market assay [
PubMed: 16800955]
- 227.
Yap
SH
, WillemsM, Van den OordJ, HabetsW, MiddeldorpJM, HellingsJA, et al. Detection of hepatitis C virus antigen by immuno-histochemical staining: a histological marker of hepatitis C virus infection. J Hepatol. 1994;20(2):275–81.
Non-blood specimens [
PubMed: 7516360]
- 228.
Yokosuka
O
, KawaiS, SuzukiY, FukaiK, ImazekiF, KandaT, et al. Evaluation of clinical usefulness of second-generation HCV core antigen assay: comparison with COBAS AMPLICOR HCV MONITOR assay version 2.0. Liver Int. 2005;25(6):1136–41.
Non-commercial or off-market assay [
PubMed: 16343063]
- 229.
You
E
, LeeW, LeeM, KimM, KangS. Comparison of HCV RNA with the positive pattern of RIBA. J Mol Diagn. 2013;15(6):888. Abstract or poster
- 230.
Yousaf
MZ
, IdreesM, SaleemZ, RehmanIU, AliM. Expression of core antigen of HCV genotype 3a and its evaluation as screening agent for HCV infection in Pakistan. Virol J. 2011;8.
Noncommercial or off-market assay [
PMC free article: PMC3152539] [
PubMed: 21787436]
- 231.
Zhang
C
, ZhouY, WangC. Detection of hepatitis C virus RNA and C33c antigen in the liver tissue from hepatitis C virus infection patients with chronic liver disease. Zhonghua nei ke za zhi. 1995;34(3):176–9.
Non-blood specimens [
PubMed: 7648939]


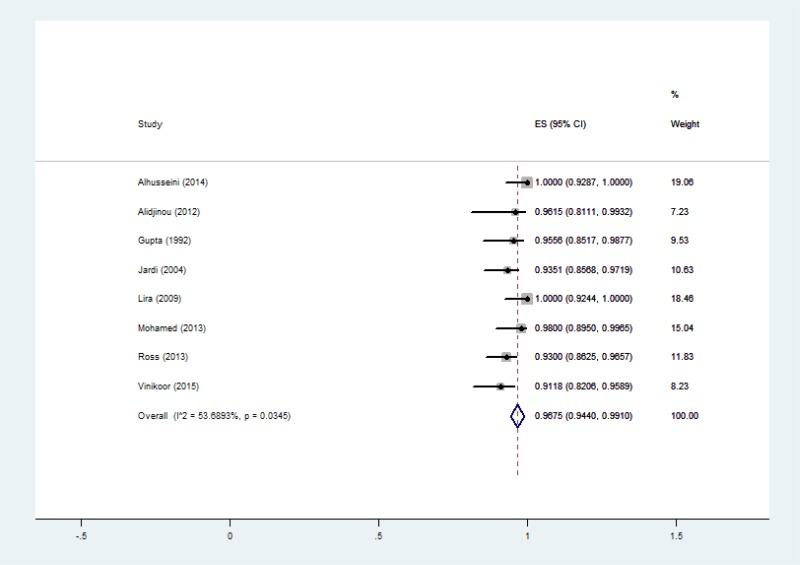
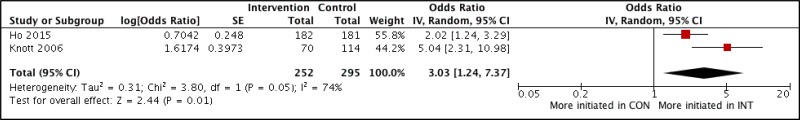
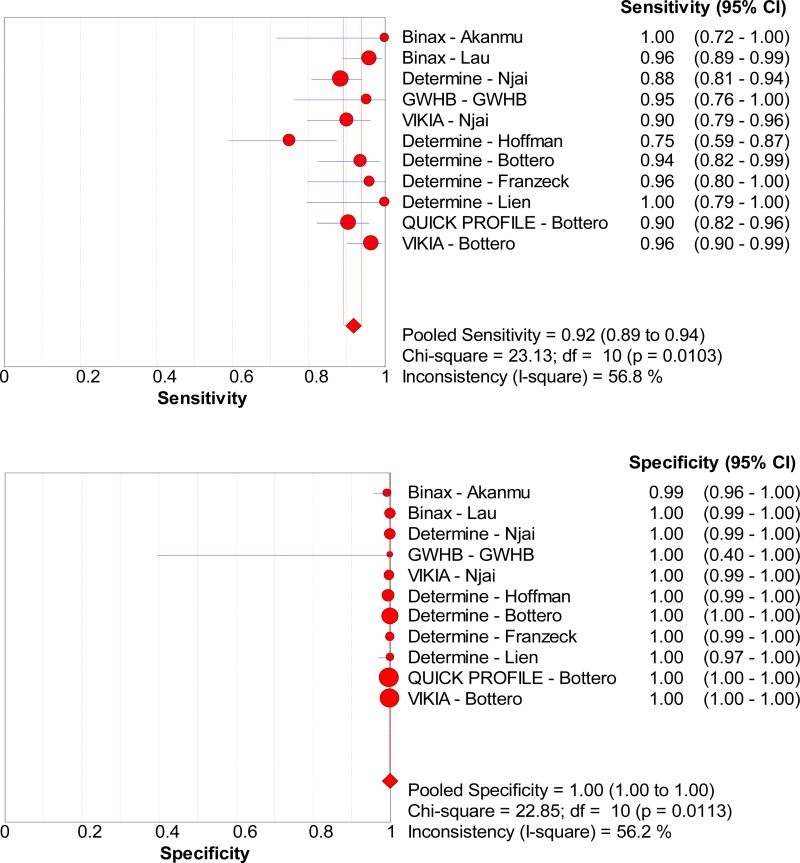
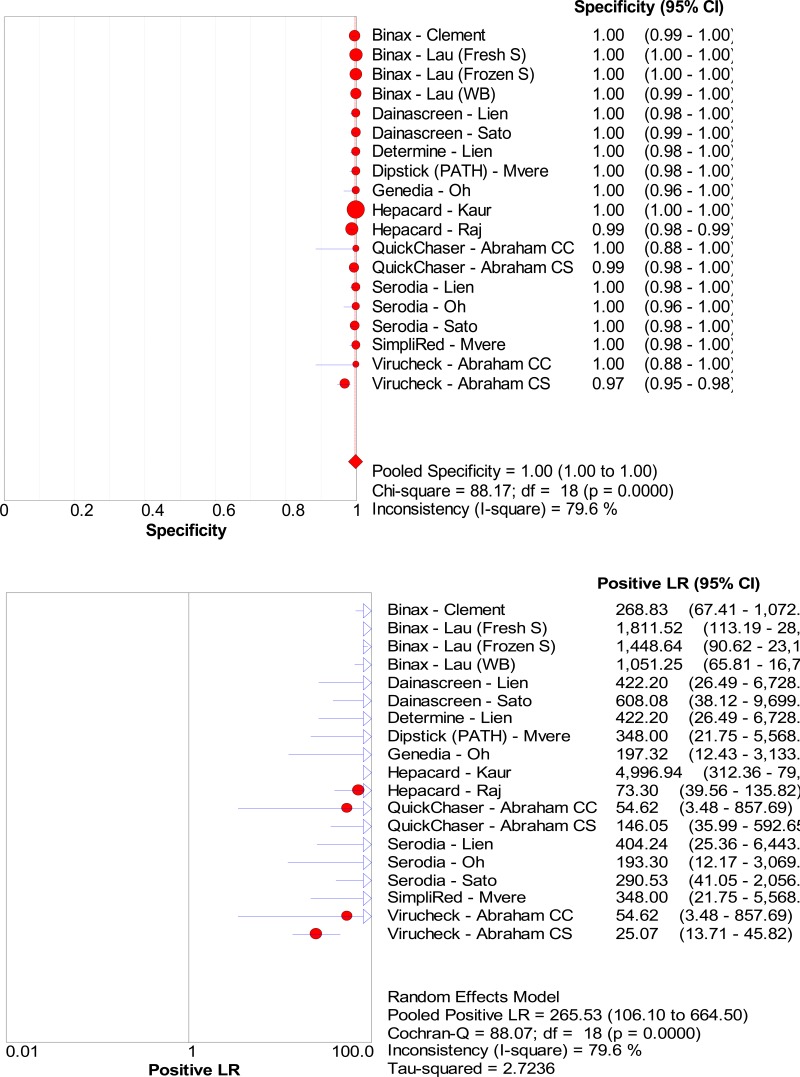
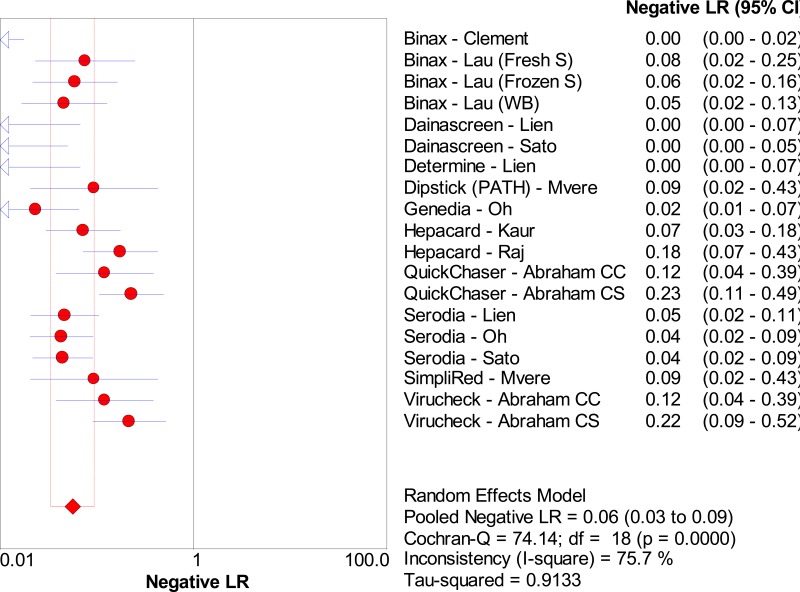
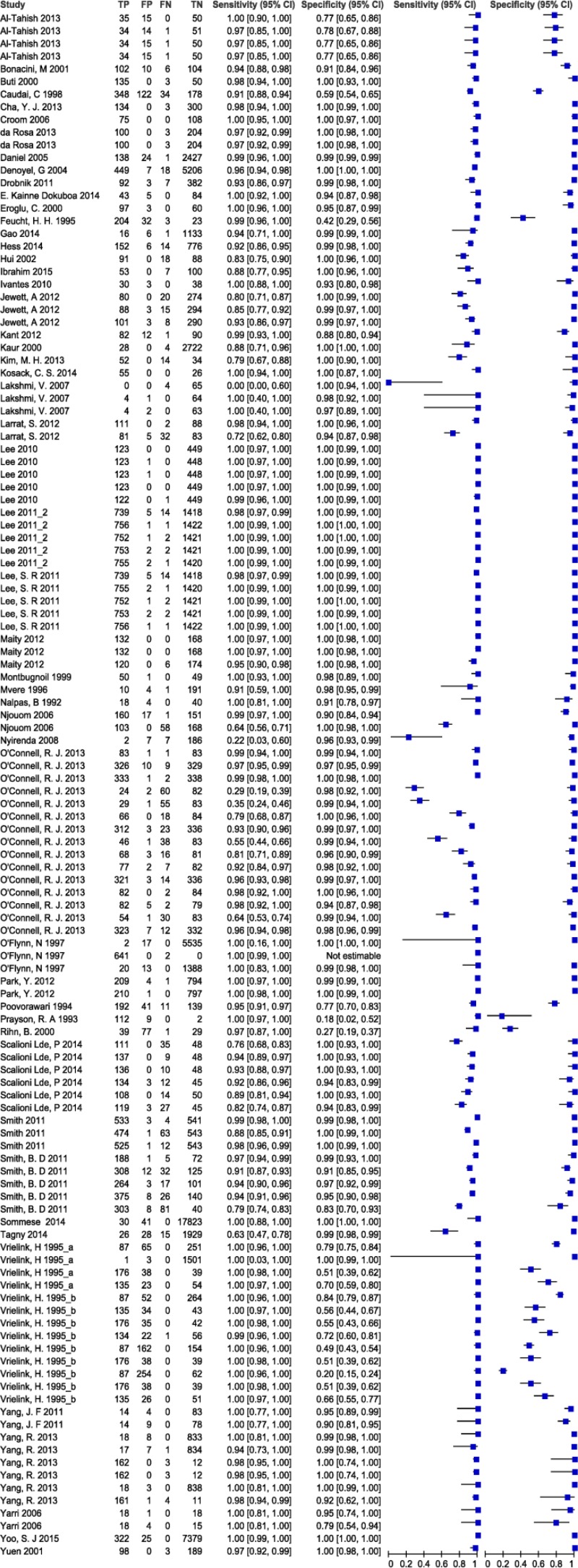
![Click on image to zoom Fig. 3. Forest plots, RDT vs EIA, ordered by [Test, Author]*.](/books/NBK442287/bin/annex5f3.jpg)
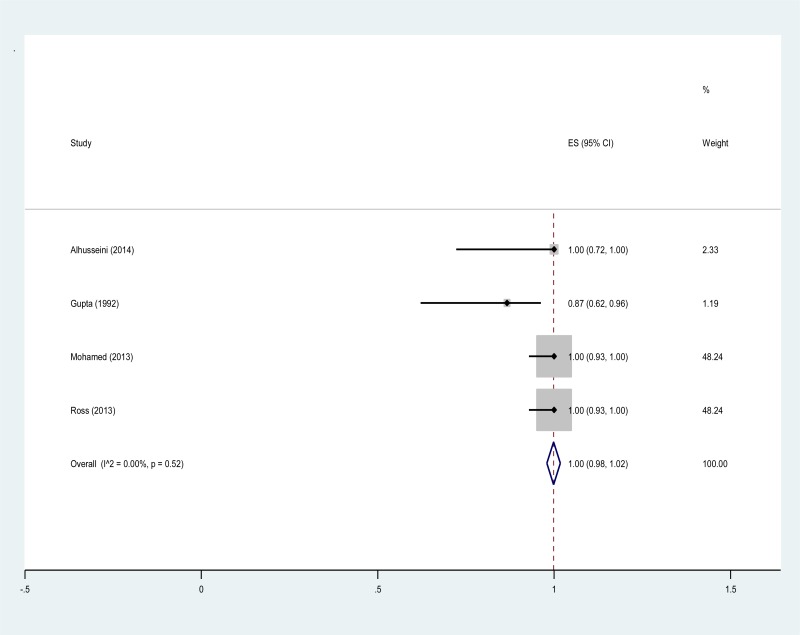
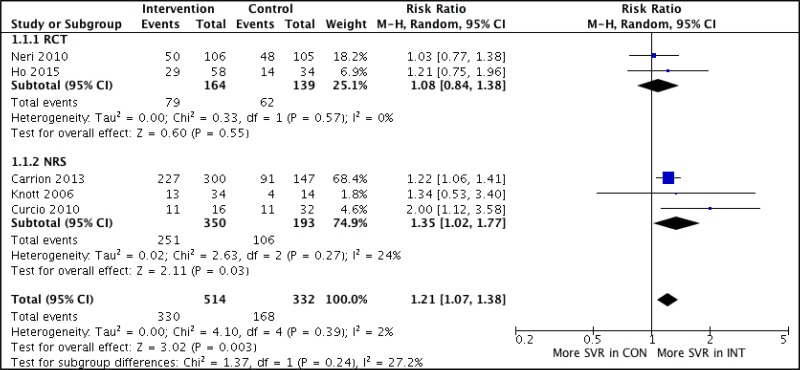
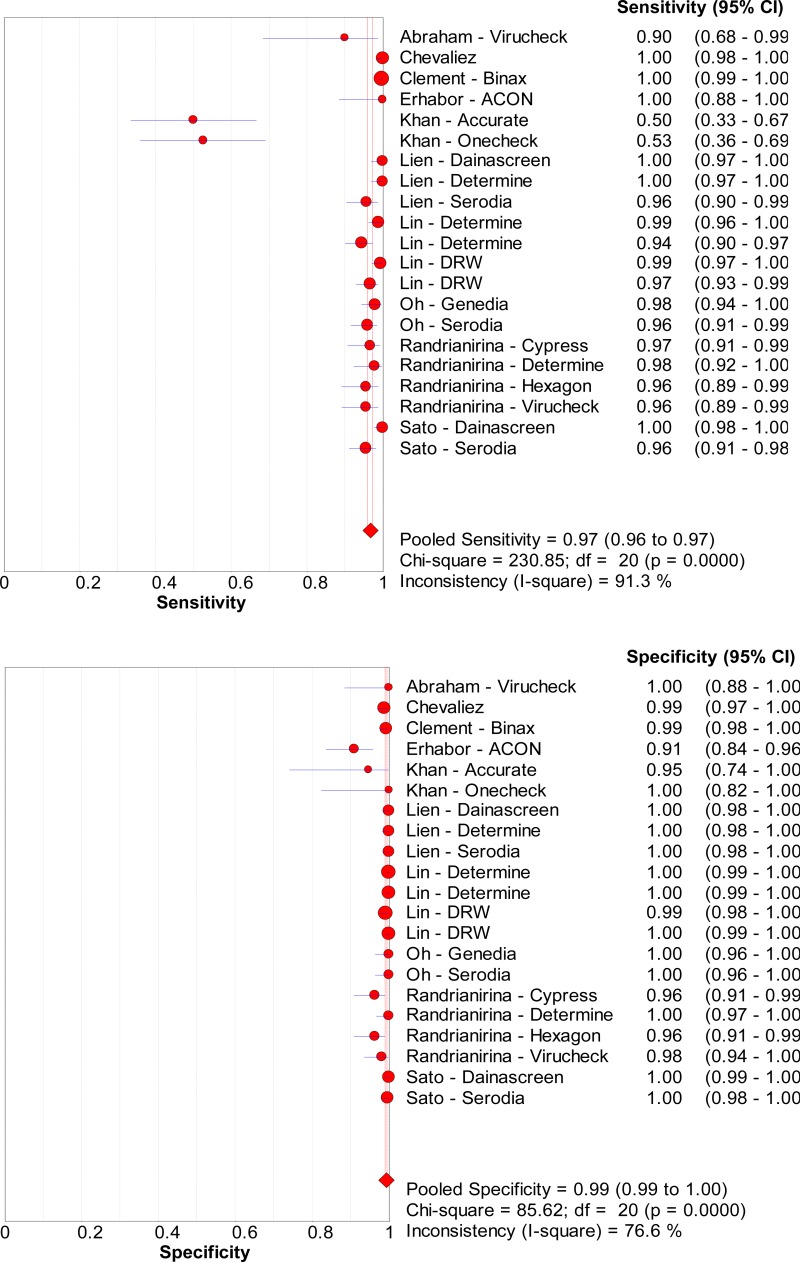
![Click on image to zoom Fig. 3. Forest plots, RDT vs EIA, ordered by [Test, Author]*.](/books/NBK442287/bin/annex5f4.jpg)
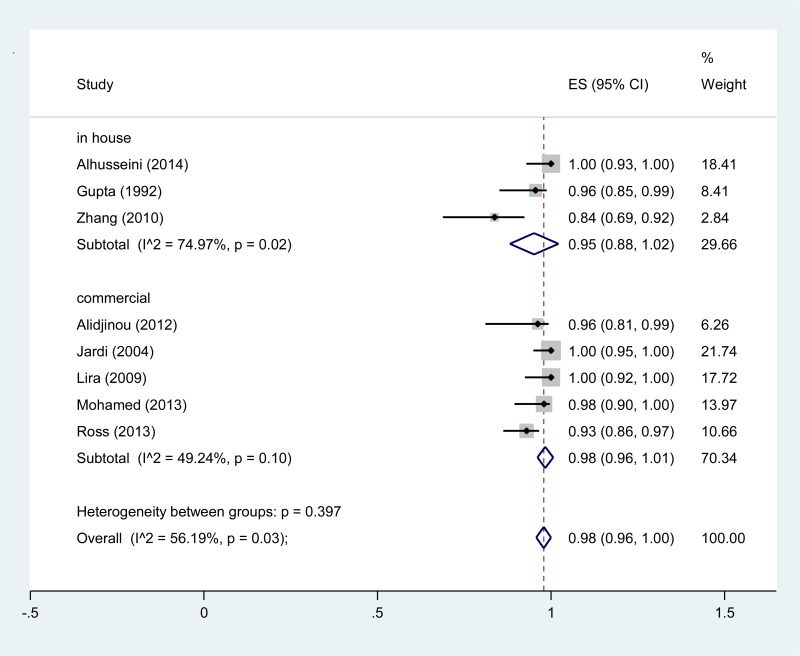
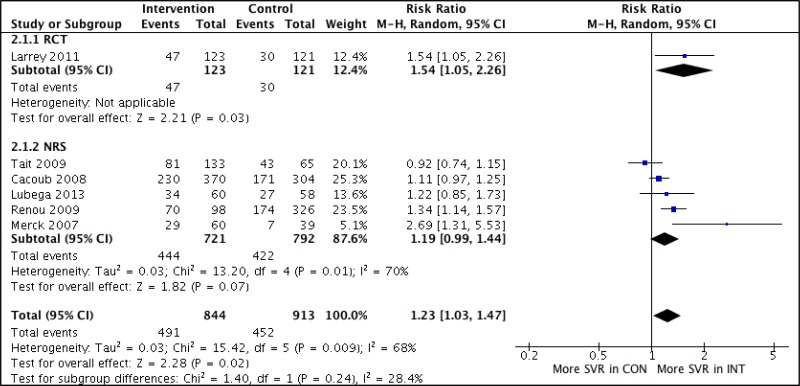
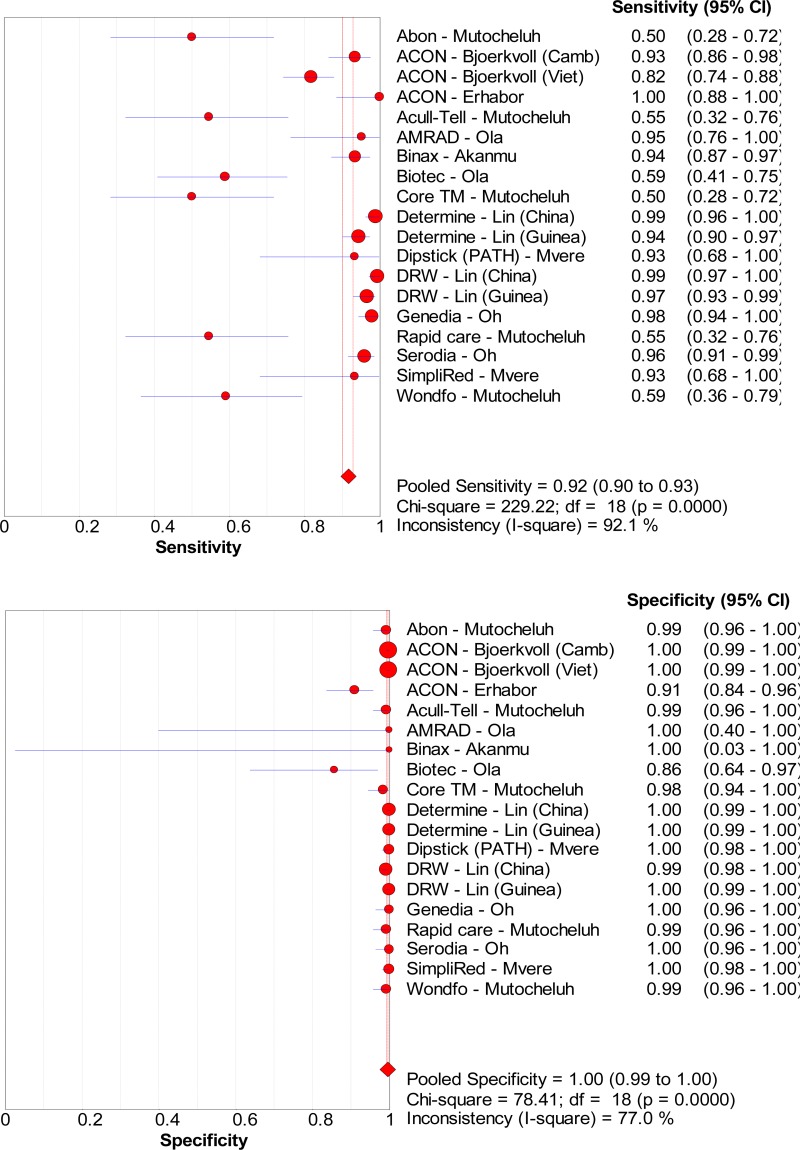
![Click on image to zoom Fig. 3. Forest plots, RDT vs EIA, ordered by [Test, Author]*.](/books/NBK442287/bin/annex5f5.jpg)
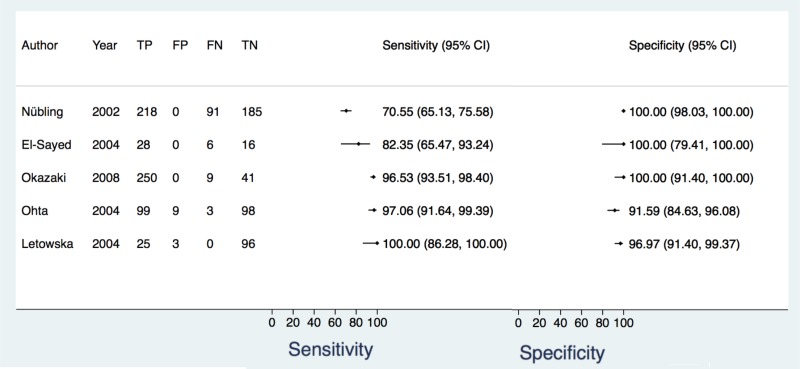
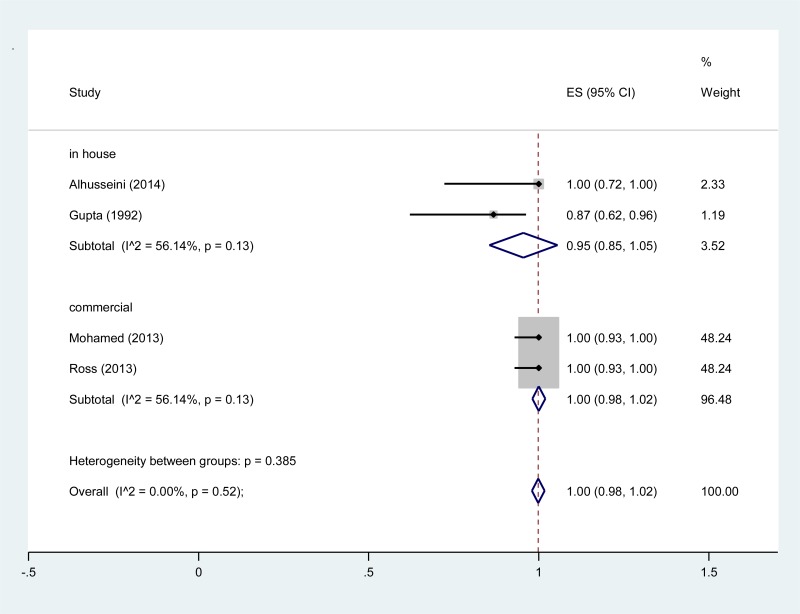
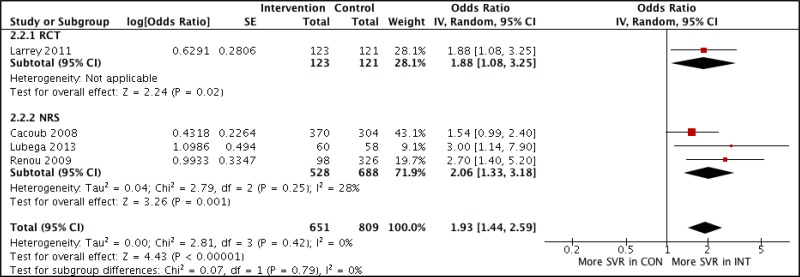
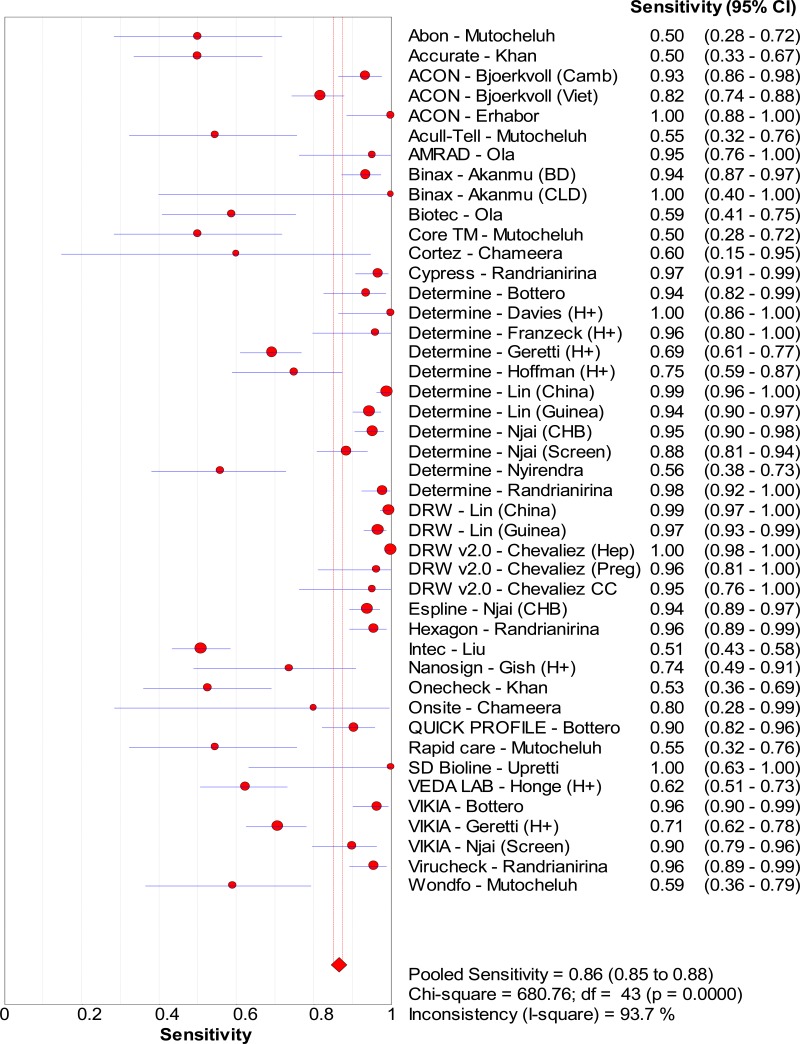
![Click on image to zoom Fig. 4. Forest plots, EIA vs EIA, ordered by [Test, Author]**.](/books/NBK442287/bin/annex5f6.jpg)
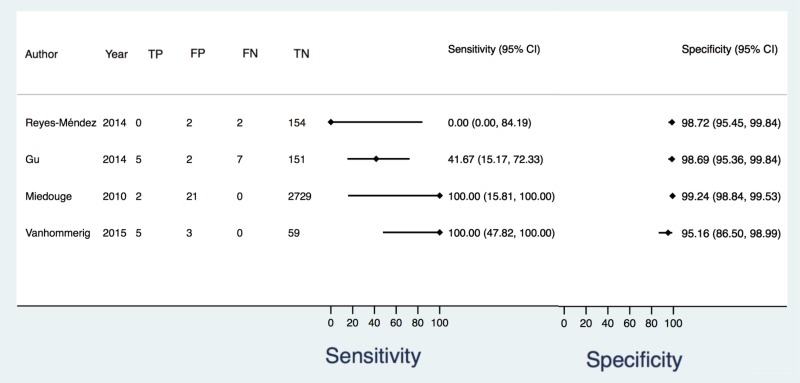
![Click on image to zoom Fig. 5. Forest plots, RDT vs NAT, ordered by [Test, Author].](/books/NBK442287/bin/annex5f8.jpg)
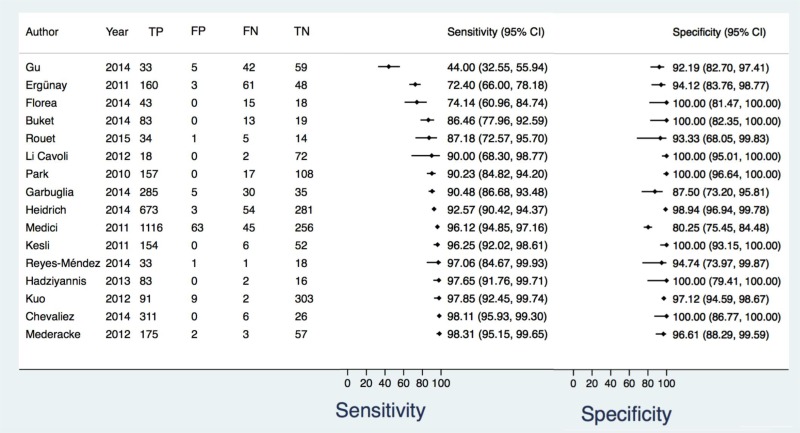
![Click on image to zoom Fig. 6. Forest plots, EIA vs NAT, ordered by [Test, Author].](/books/NBK442287/bin/annex5f9.jpg)
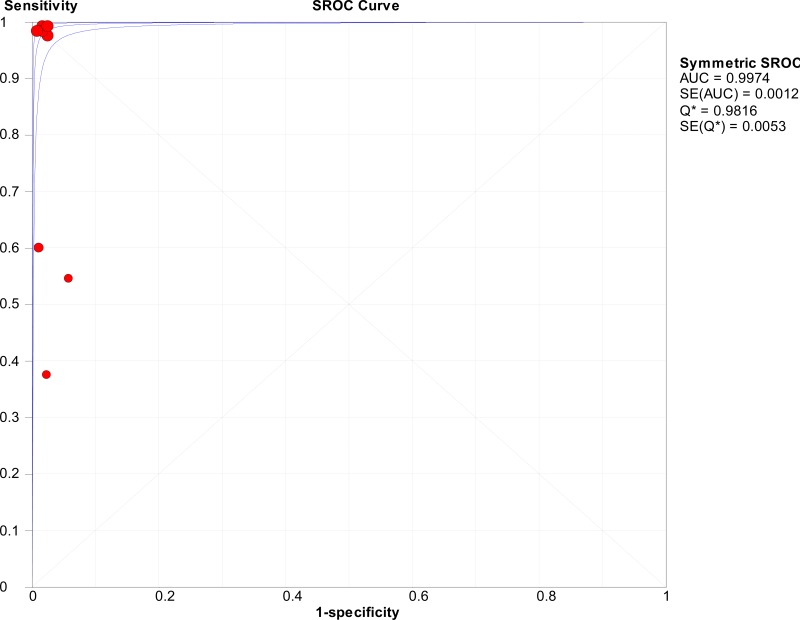
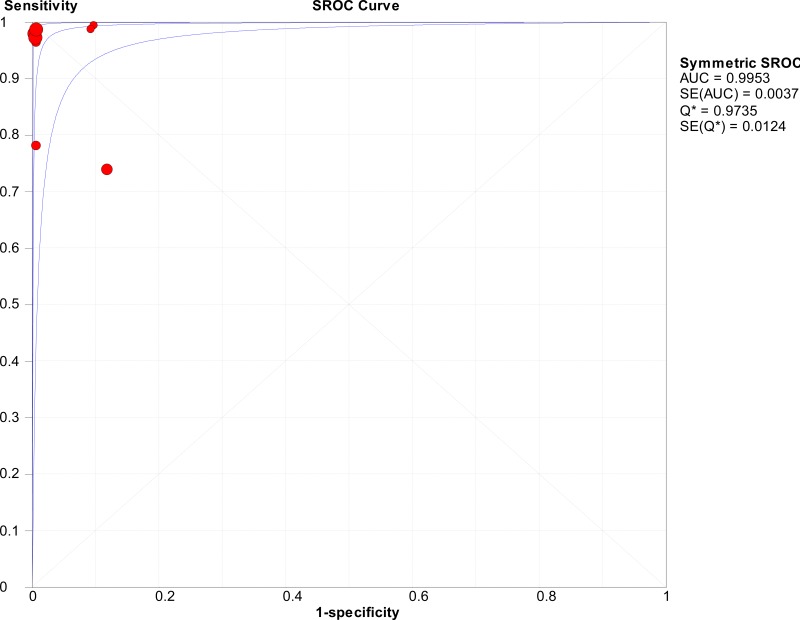
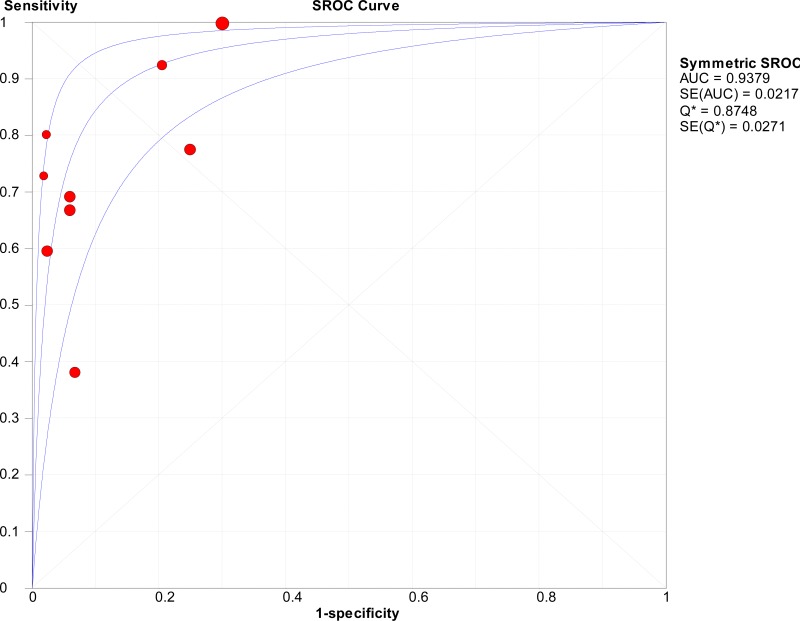
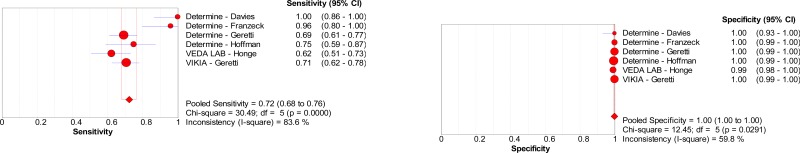
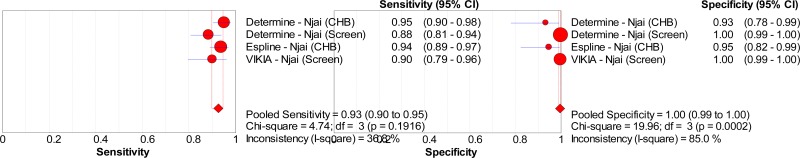
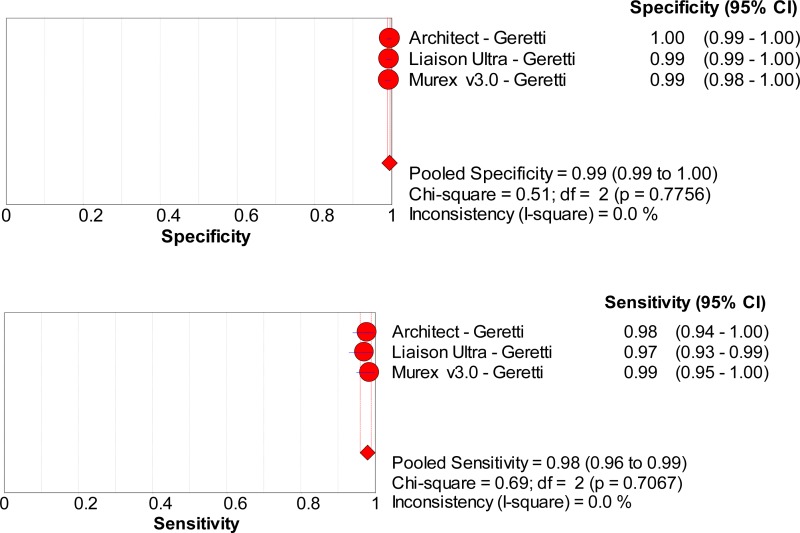

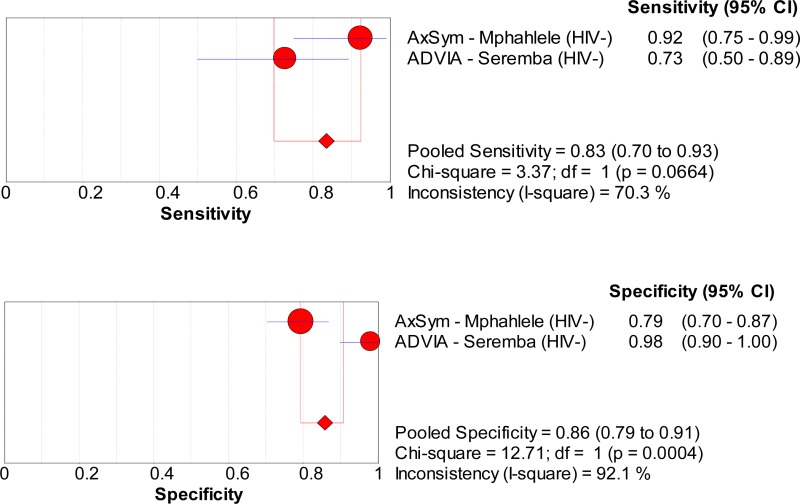
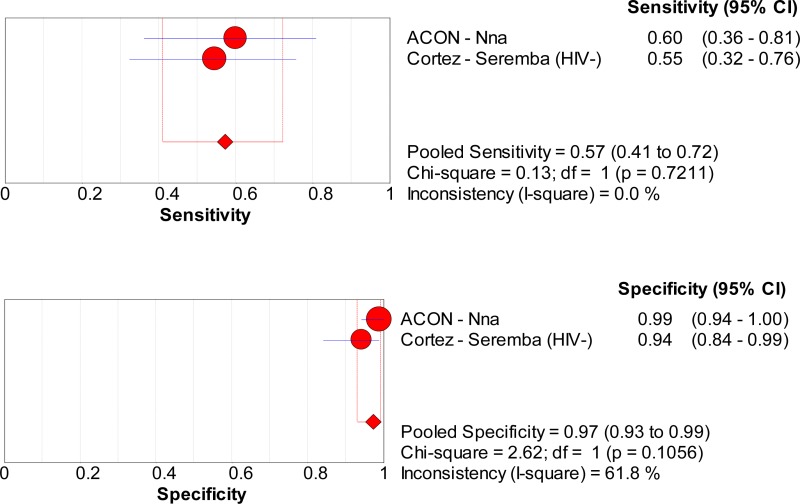
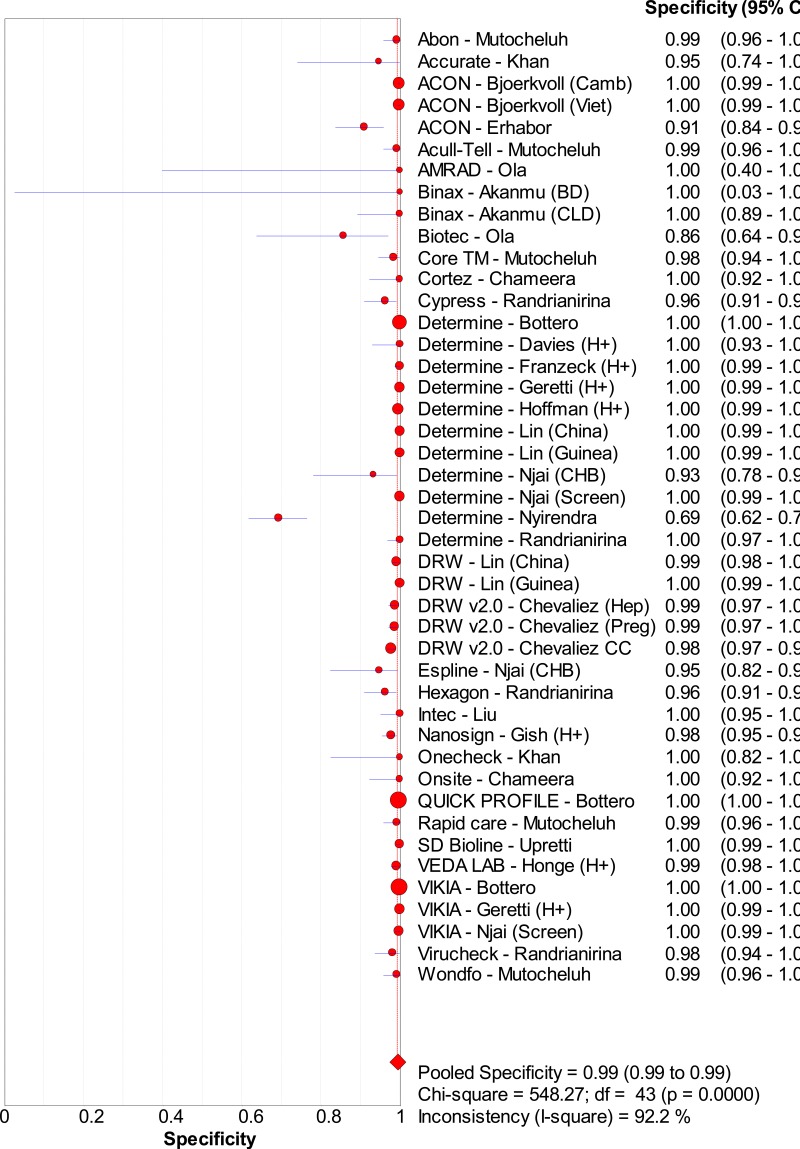
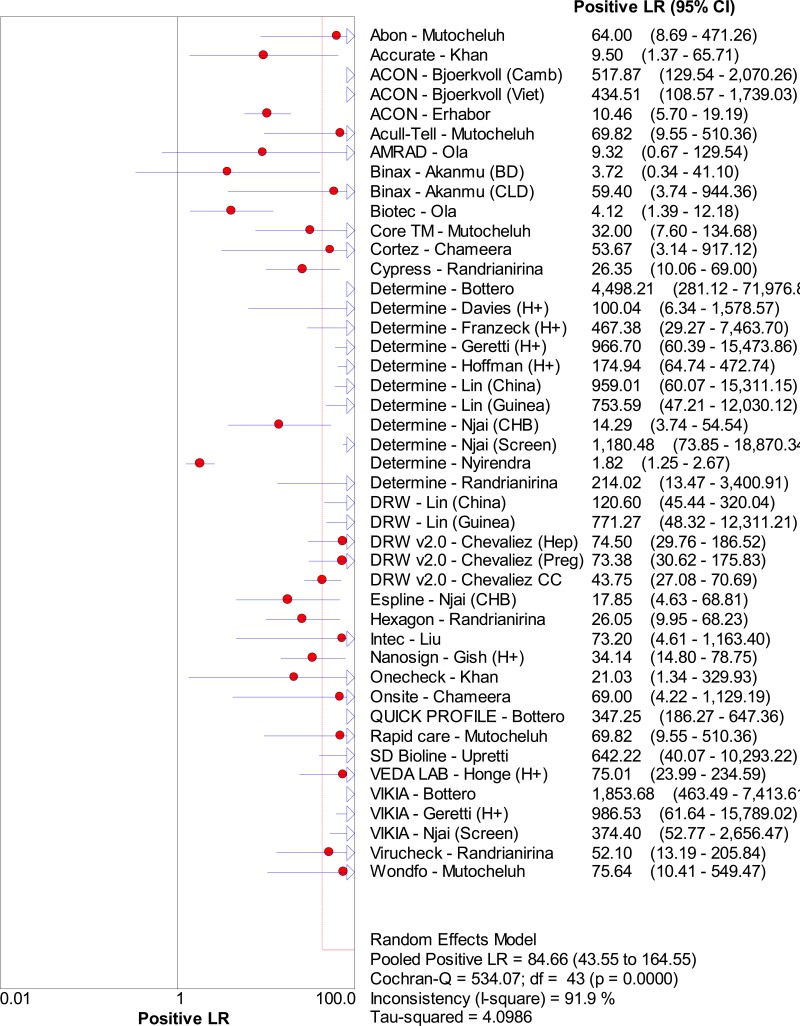
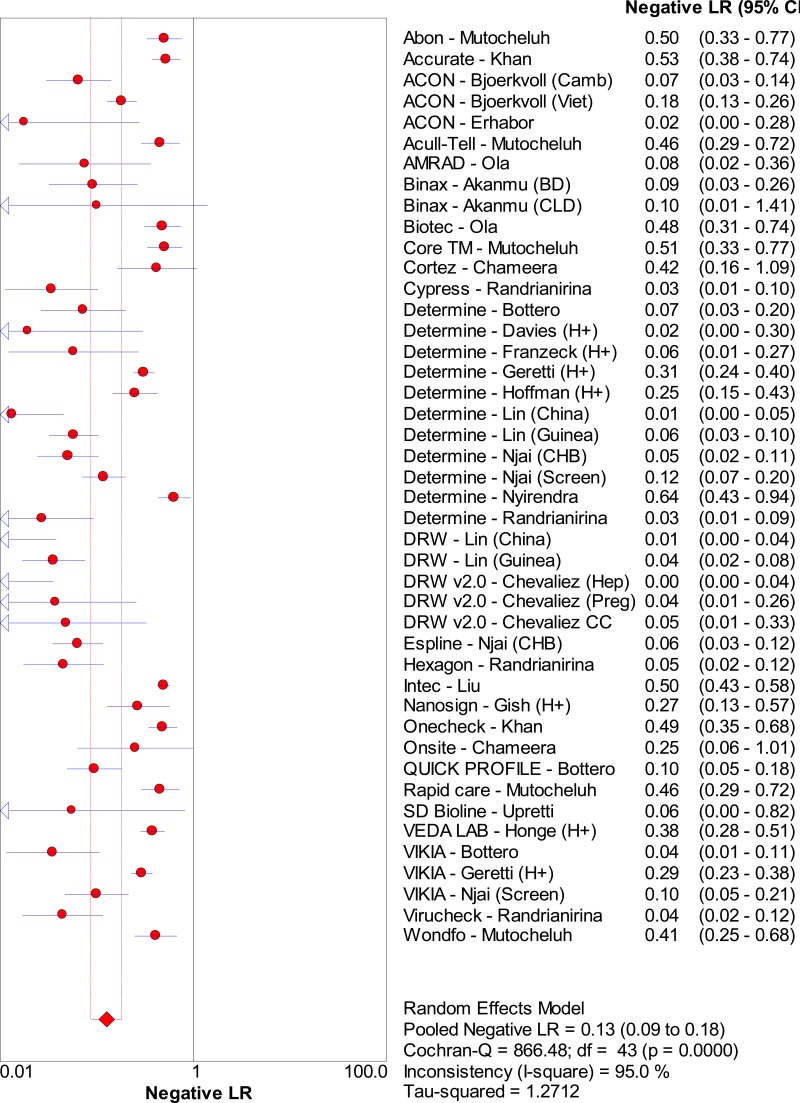
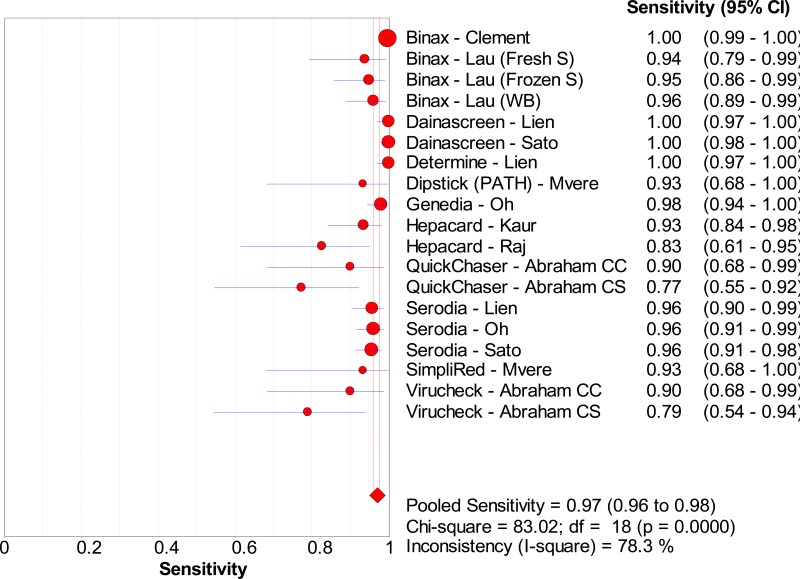
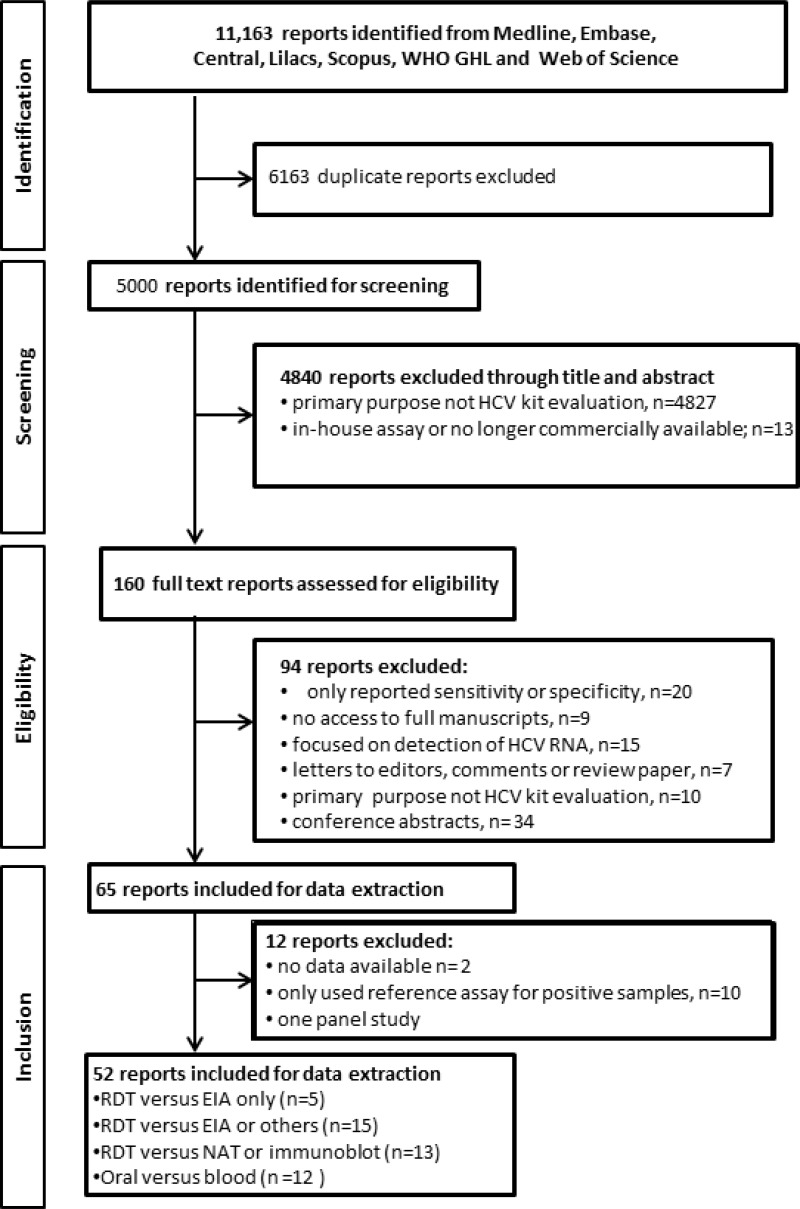
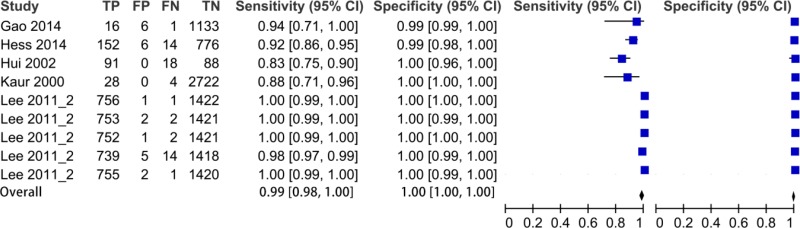
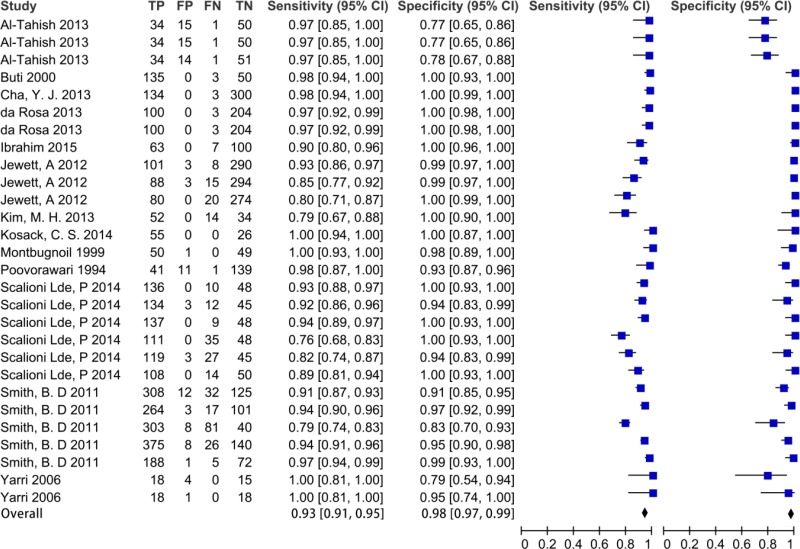
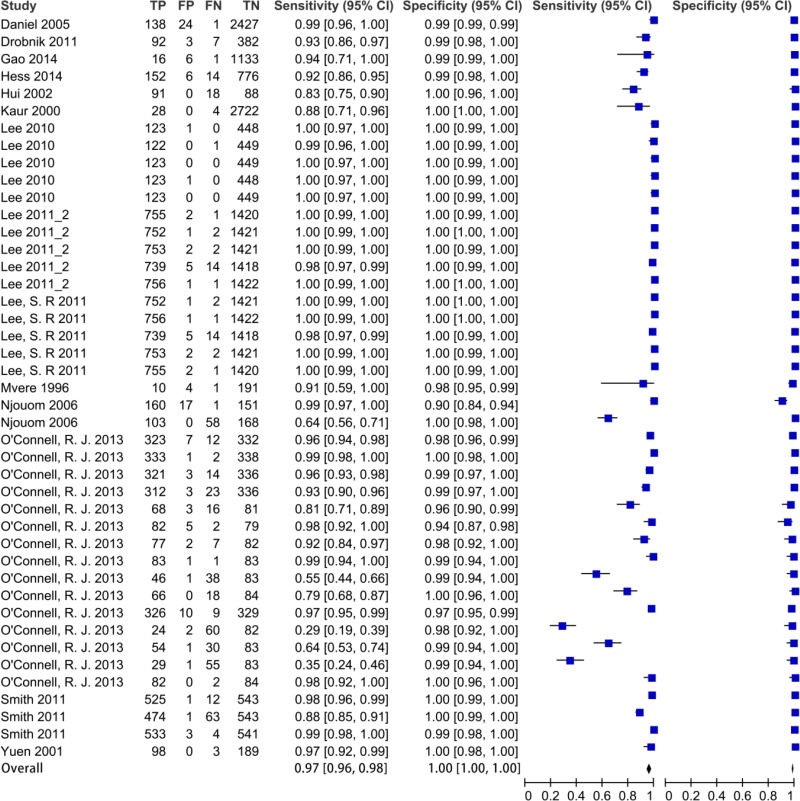
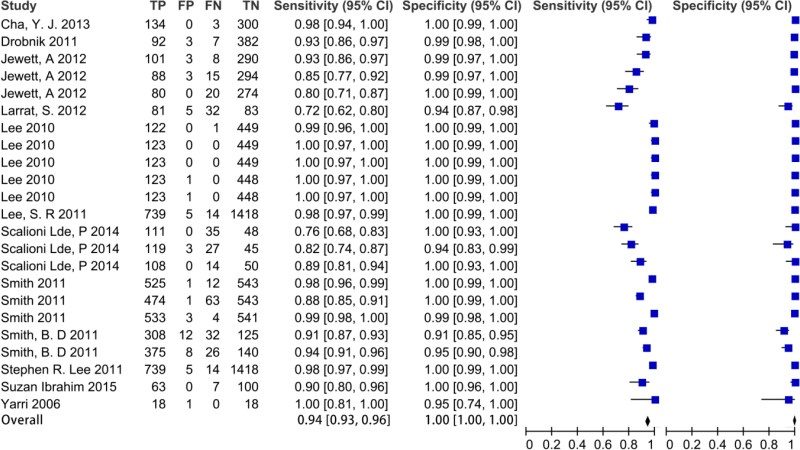
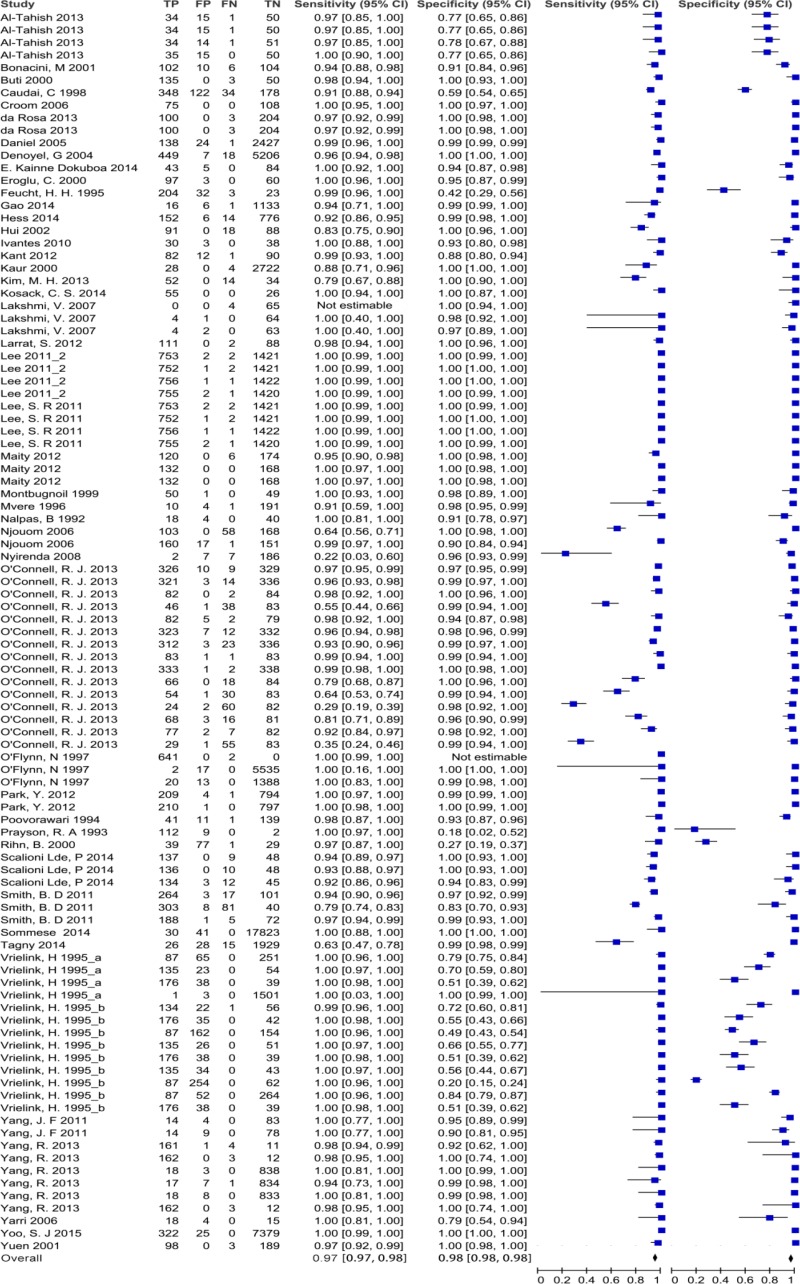
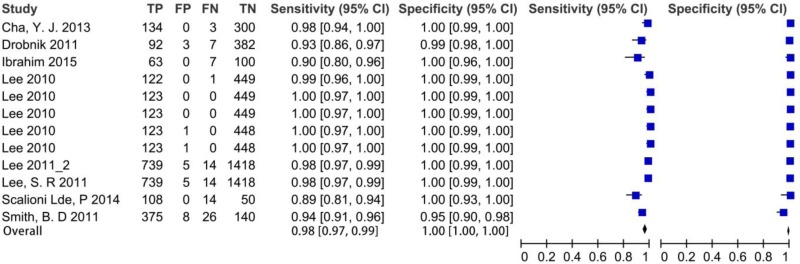
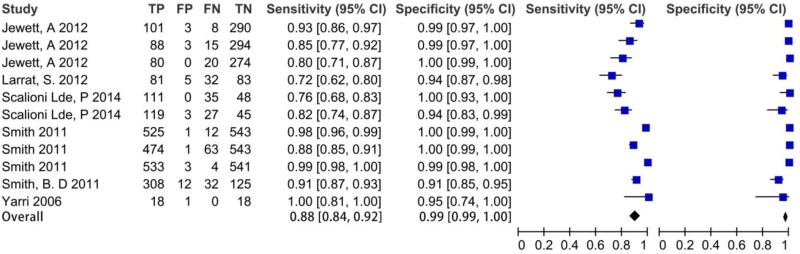
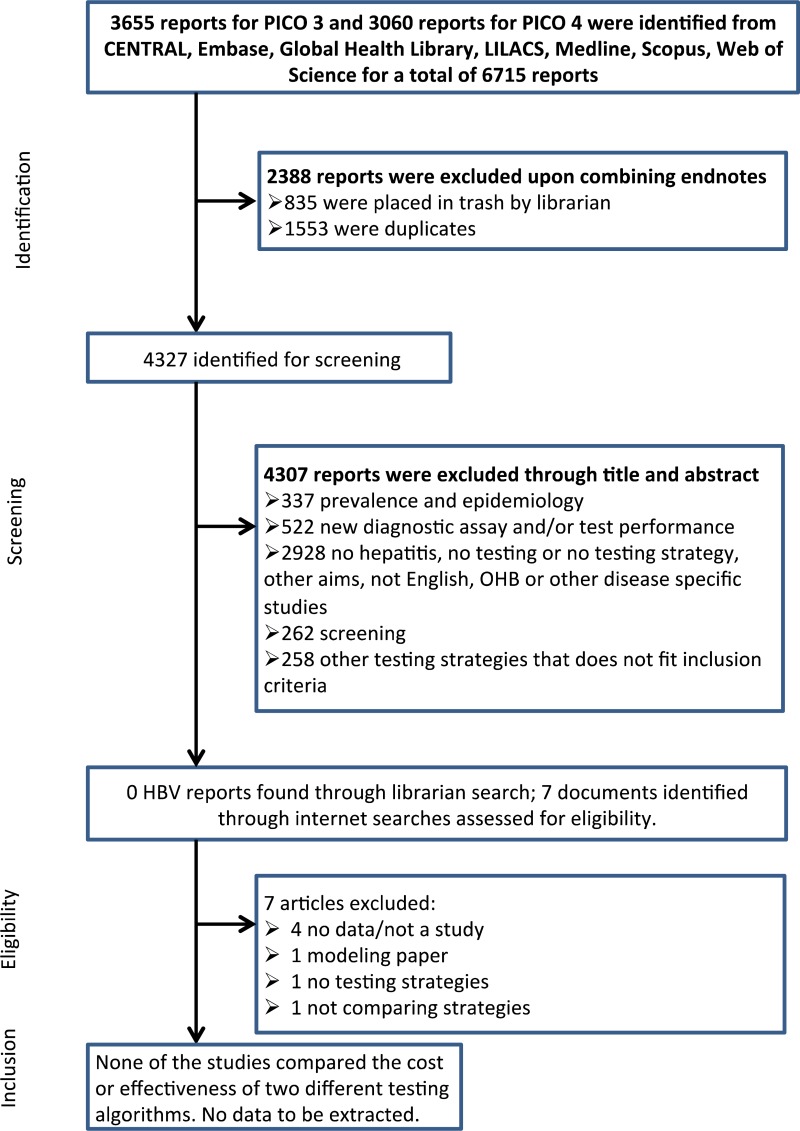
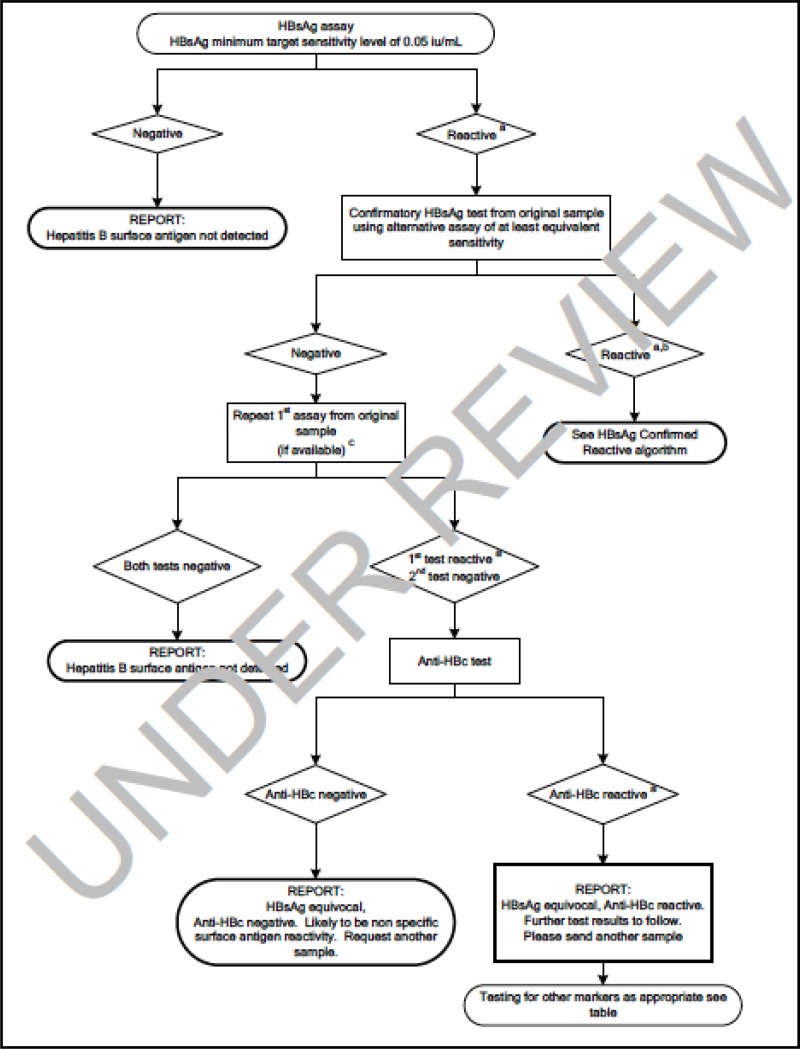
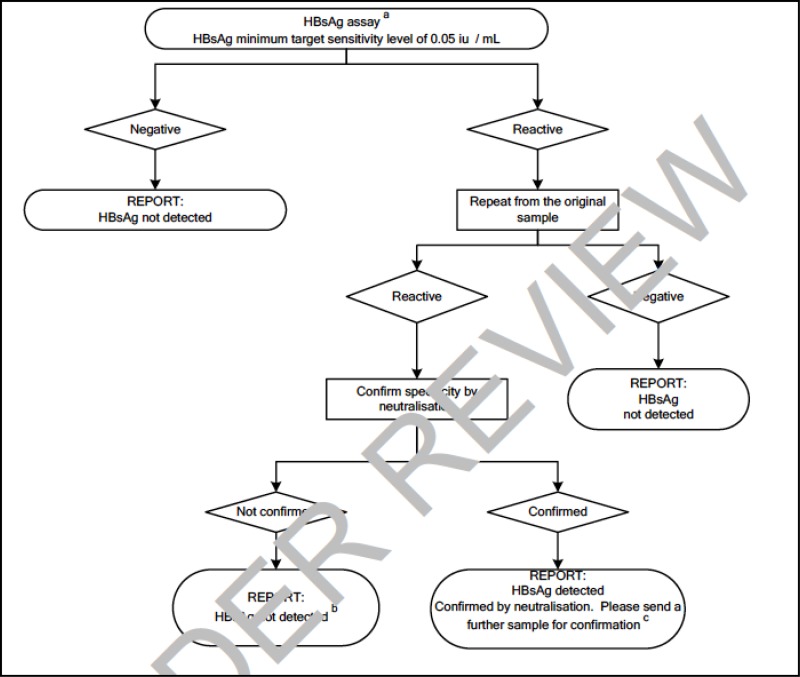
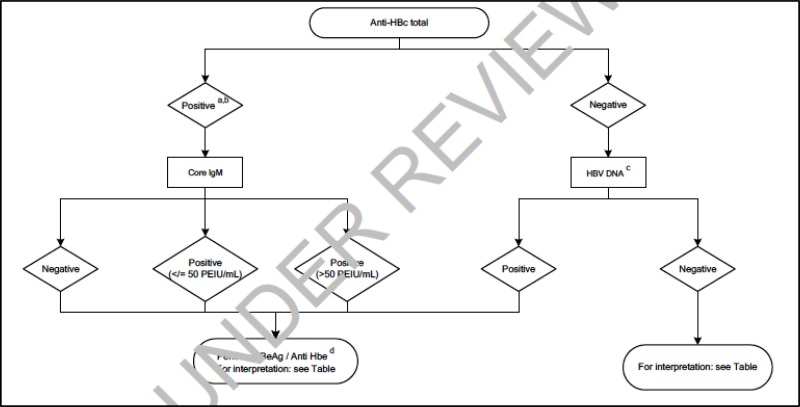
![Click on image to zoom Fig. 1. Options for HBV screening, which may include HBsAg in a one-test strategy (e.g. a single HBsAg using a rapid diagnostic test [RDT] or enzyme immunoassay [EIA]) and two-test strategies (second RDT or EIA or neutralization with EIA).](/books/NBK442287/bin/annex5f39.jpg)
![Click on image to zoom Fig. 1. Options for HBV screening, which may include HBsAg in a one-test strategy (e.g. a single HBsAg using a rapid diagnostic test [RDT] or enzyme immunoassay [EIA]) and two-test strategies (second RDT or EIA or neutralization with EIA).](/books/NBK442287/bin/annex5f40.jpg)
![Click on image to zoom Fig. 1. Options for HBV screening, which may include HBsAg in a one-test strategy (e.g. a single HBsAg using a rapid diagnostic test [RDT] or enzyme immunoassay [EIA]) and two-test strategies (second RDT or EIA or neutralization with EIA).](/books/NBK442287/bin/annex5f41.jpg)