1. Background
Currently, the World Health Organization (WHO) estimates that chronic hepatitis B virus (HBV) infection affects close to 260 million persons and causes an estimated 900 000 deaths annually through manifestations of chronic liver disease, such as cirrhosis or hepatocellular carcinoma (HCC). The regions with the highest prevalence of chronic HBV infection (CHB) are the Western Pacific and Africa (1). In 2016, the World Health Assembly endorsed the Global Health Sector Strategy (GHSS) on viral hepatitis, which calls for an elimination of HBV worldwide as a public health threat by 2030, to be accomplished through reducing the incidence of CHB by 90%, and its mortality by 65% (2).
Chronic infection is more likely to develop when HBV is acquired early in life, and therefore, perinatal mother-to-child transmission (MTCT) is a major contributor to the incidence of CHB (3). Moreover, the risk of developing chronic liver disease, including HCC, may be higher in those who acquired CHB through MTCT compared to those who ended up with CHB through horizontal transmission later in life (4,5).
In order to achieve the WHO’s global hepatitis elimination plan, it is imperative to prevent MTCT of HBV (6). Since 2009, WHO makes a universal recommendation to administer timely hepatitis B vaccines to all newborn babies within 24 hours of birth to prevent MTCT and early childhood transmission (7). Although the birth dose vaccines alone should be enough to prevent MTCT from mothers with CHB who have low HBV viral replication (8,9), 20–30% of women with high viral load infect their newborns despite timely birth dose vaccination (9,10). Therefore, in resource-rich countries, pregnant women are screened for hepatitis B surface antigen (HBsAg), and subsequently for hepatitis B e antigen (HBeAg), to identify high-risk infants who would benefit from hepatitis B immunoglobulin (HBIG) in addition to timely birth dose vaccination (11). However, despite this active and passive immunoprophylaxis, a substantial proportion of infants are still infected when their mothers have a very high viral load, particularly when the serum HBV DNA level exceeds 200 000 IU/mL (12). Consequently, in high-income countries, HBV DNA quantification has become a part of antenatal HBV testing to identify highly viraemic women who have a residual risk of MTCT despite administration of both hepatitis B vaccine and HBIG to neonates at birth (12–14), and who thus require antiviral therapy during pregnancy for minimizing its risk (15).
However, in low- and middle-income countries, such additional measures to prevent MTCT have rarely been implemented (16). Following antenatal screening for HBsAg, it is essential to quantify serum HBV DNA levels using the nucleic acid test (NAT) to decide whom to treat and not to treat during pregnancy to prevent MTCT. However, access to NAT is severely limited in these countries. The current standard NAT assay, which is real-time polymerase chain reaction (PCR), is hardly accessible due to its high cost (US$ 60–200/assay) and its need for a sophisticated laboratory with highly skilled laboratory staff (1). Alternatively, detection of HBeAg using laboratory-based immunoassays, such as enzyme immunoassay (EIA) and chemiluminescence immunoassay (CLIA), or rapid diagnostic test (RDT) with lateral flow immunochromatographic assay, may largely overcome these limitations, because these tests may be more readily available and affordable (US$ 1–30/assay) than HBV DNA NAT in such settings (17).
3. Methods
3.1. Narrative review question
Can HBeAg test be used instead of NAT to diagnose high HBV DNA levels in order to assess eligibility for antiviral therapy initiation in pregnant women with CHB to prevent MTCT?
3.2. PICO questions
We obtained evidence to answer the following questions ().
Table 1PICO questions for this systematic review
View in own window
| PICO2A | PICO2B | PICO2C |
|---|
| What is the performance of HBeAg to diagnose high HBV DNA levels in pregnant women with CHB? | What is the performance of HBeAg in pregnant women with CHB to predict the risk of MTCT? | What is the performance of different HBV DNA thresholds in pregnant women with CHB to predict the risk of MTCT? |
|---|
| Population | Pregnant women with CHB*1 without concomitant anti-HBV therapy | Same as PICO2A | Same as PICO2A |
|---|
| Intervention | Maternal HBeAg test during pregnancy*2 | Same as PICO2A | Maternal HBV DNA levels during pregnancy*4
Dichotomized into high and low using the following thresholds: ≥20,000, 10^5, 10^6, 10^7 and 10^8 IU/mL
|
|---|
| Comparison | Maternal HBV DNA levels during pregnancy*3,*4
Dichotomized into high and low using the following thresholds: ≥20 000, 10^5, 10^6, 10^7, & 10^8 IU/mL
| MTCT defined as:
| Same as PICO2B |
|---|
| Outcomes | Sensitivity and specificity of HBeAg tests*5 to diagnose each of different HBV DNA thresholds | Sensitivity and specificity of HBeAg tests to predict MTCT*6 | Sensitivity and specificity of HBV DNA test to predict MTCT*6 |
|---|
- *1
CHB was defined as HBsAg seropositivity on two occasions at least 6 months apart. However, because new HBV infections in adults are uncommon in highly endemic areas where the vast majority of HBsAg-positive people acquired the infection perinatally or during childhood, HBsAg positivity on only one occasion (at antenatal care) in women living in highly prevalent countries was assumed to reflect CHB (21).
- *2
Maternal HBeAg test performed after child delivery was not considered. The test result should be reported positive or negative; an indeterminate result was not considered for the meta-analysis. Instead, the frequency of the indeterminate result in each study was extracted and reported. The following HBeAg immunoassays were considered:
Lateral flow immunochromatographic rapid diagnostic test (RDT)
Enzyme immunoassay (EIA)
Chemiluminescence immunoassay (CLIA)
Radioimmunoassay (RIA)
Counting immunoassay (CIA)
Fluoroimmunoassay (FIA)
- *3
It is ideal to have both HBeAg and HBV DNA measurements from the same sample, or at least from a sampling done at the same time. However, a study was still considered whenever both markers were measured during the same period of pregnancy, even if they were not measured using samples collected on the same day.
- *4
There are two types of NAT: qualitative (undetectable or detectable) and quantitative. When NAT provided a continuous value through quantification of HBV DNA levels, the value was dichotomized into high and low according to a threshold used in each included study. In order to have a wide range of estimations, the HBV DNA threshold used in each included study to dichotomize HBV DNA levels into high and low should have been greater than or equal to 20 000 IU/mL. Similarly, when NAT provided only a qualitative binary result (detectable or undetectable), the limit of detection of the qualitative NAT should have been greater than or equal to 20 000 IU/mL.
- *5
For example, in the case of HBV DNA levels of ≥200 000 IU/mL, the sensitivity and specificity were defined as below:
- ▪
- ▪
In order to have these estimates, a study needed to provide sufficient data for us to draw a 2x2 or 2x1 table with the cross-classification of the reference test results (high vs low HBV DNA levels) and the index test results (positive vs negative HBeAg serostatus).
- *6
The outcome was stratified by the type of preventive measures: timely birth dose vaccine (yes or no); and HBIG at birth (yes or no). We only considered studies in which sensitivity and specificity estimates could be stratified by the type of preventive measure provided to the mother–child pairs. We did not consider studies that provided antiviral therapy to mothers during pregnancy, since our objective is to evaluate these HBV markers as a tool to identify pregnant women who would benefit from antiviral therapy during pregnancy.
Other inclusion and exclusion criteria: study design, languages, dates of publication
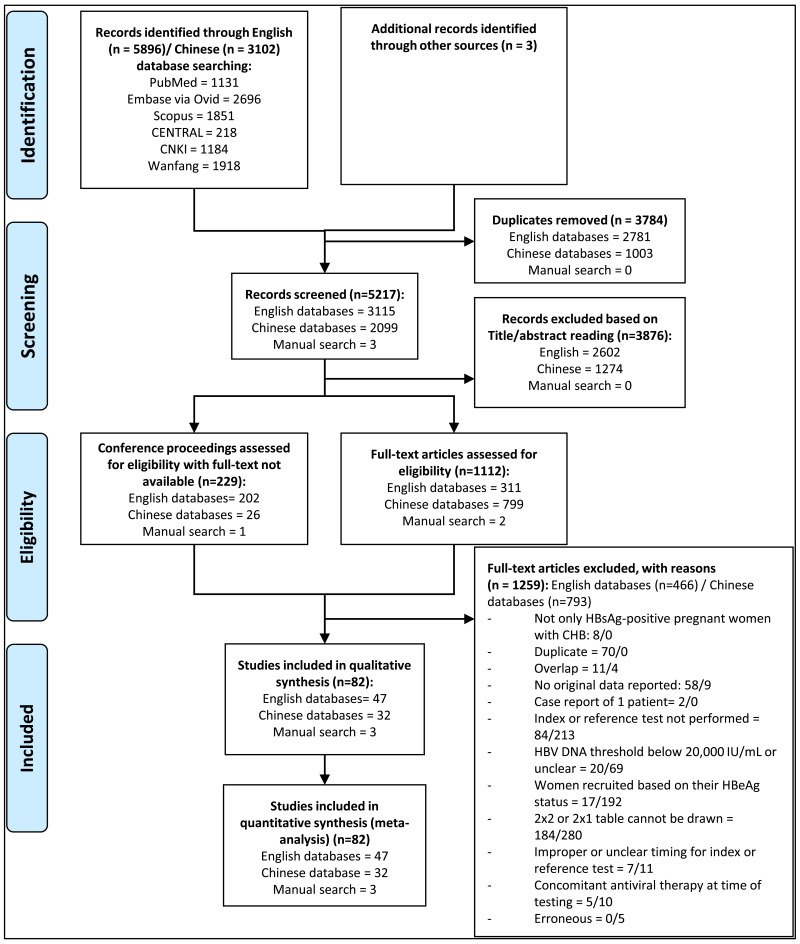
We included studies with any design, published in any language, which used an HBV DNA threshold to dichotomize HBV DNA levels into high and low. This threshold needed to be at least higher than 20 000 IU/mL. Moreover, studies needed to provide sufficient data to draw a 2x1 or 2x2 table with the cross-classification of the reference test results (high vs low HBV DNA levels) and the index test results (positive vs negative HBeAg serostatus) in pregnant women with CHB without concomitant anti-HBV therapy. We excluded studies that selected participants based on the index test (i.e. maternal HBeAg status) to avoid verification bias (22). Studies published between 1 January 2000 and 3 April 2019 were considered.
3.3. Post-hoc analyses
For the post-hoc analyses, we included studies evaluating mother–child pairs, in which child outcomes could be stratified by different maternal HBV DNA levels during pregnancy with a narrow range (≤1.0 log IU/mL; such as <4.0, 4.0–4.9, 5.0–5.9, 6.0–6.9, and ≥7.0 log IU/mL). At each stratum defined by maternal viral load, there should be ≥10 infants assessed for MTCT. We excluded studies that selected participants based on maternal HBeAg status or maternal viral load to avoid verification bias (22). Studies published between 1 January 2000 and 3 April 2019 were considered.
3.4. Search strategy
The search terms employed covered “hepatitis B infection” AND “viral load” AND “pregnancy” and their variations. The databases searched included: four English-language databases (PubMed, EMBASE, Scopus, and CENTRAL (the Cochrane Library)); and two Chinese-language databases (the China National Knowledge Infrastructure [CNKI] and the Wanfang database). The search strategies used for each of the databases are presented in Appendix A.
A manual search through the references of the included studies, as well as through those of relevant systematic reviews identified through the literature search, was undertaken to identify any further eligible studies. Expert opinion was also sought to include other relevant studies.
3.5. Conduct of the review
Titles and abstracts for all of the publications identified by the search strategy were independently screened for relevance by two reviewers (PB and KY). Following selection of potentially eligible studies, full-text reading and reviewing was independently performed. Finally, the two reviewers discussed the list of eventually eligible studies, and if discrepancies existed that could not be resolved between the two reviewers, a third reviewer (YS) was consulted in order to make the final decision. For the Chinese databases, the same procedure was followed by two independent Chinese reviewers (YL and TZ).
For all potentially eligible studies, if information was lacking within the full-text article that limited the ability to make a final decision on whether or not the study should be included, the corresponding author of that study was contacted by mail or phone.
The final protocol for this review was registered on the international prospective register of systematic reviews (PROSPERO) with the registration number: CRD42019138227 prior to starting the data analysis.
3.6. Quality appraisal
The quality of included studies was assessed independently by two reviewers.
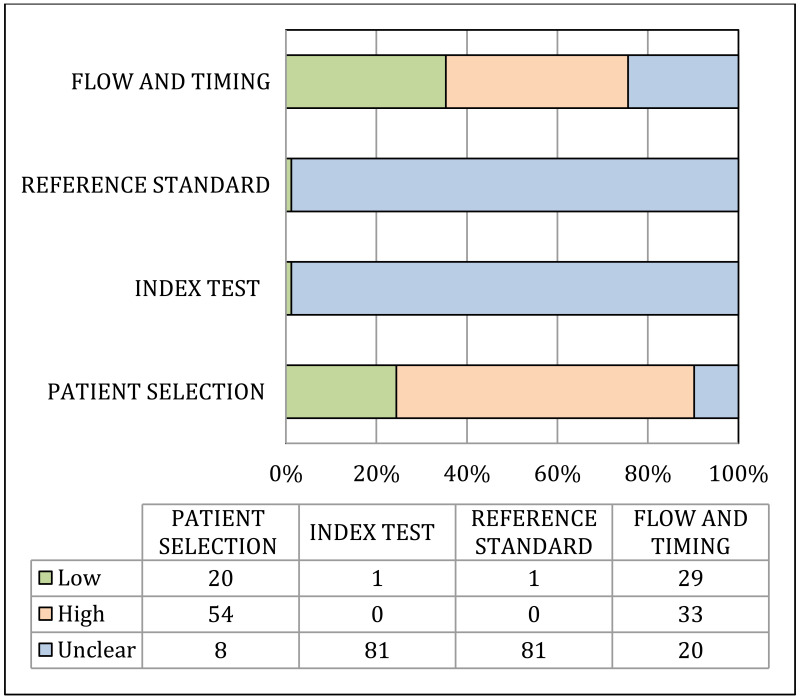
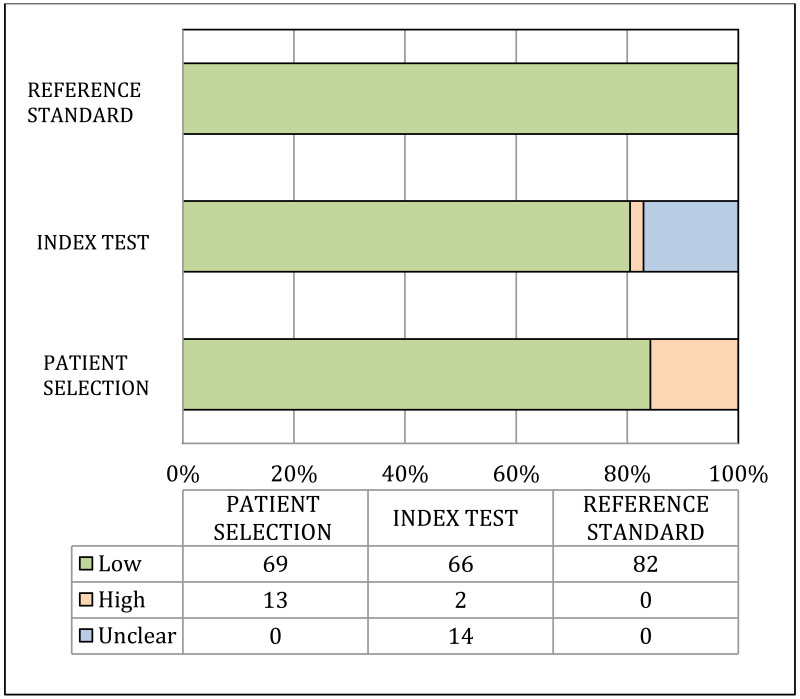
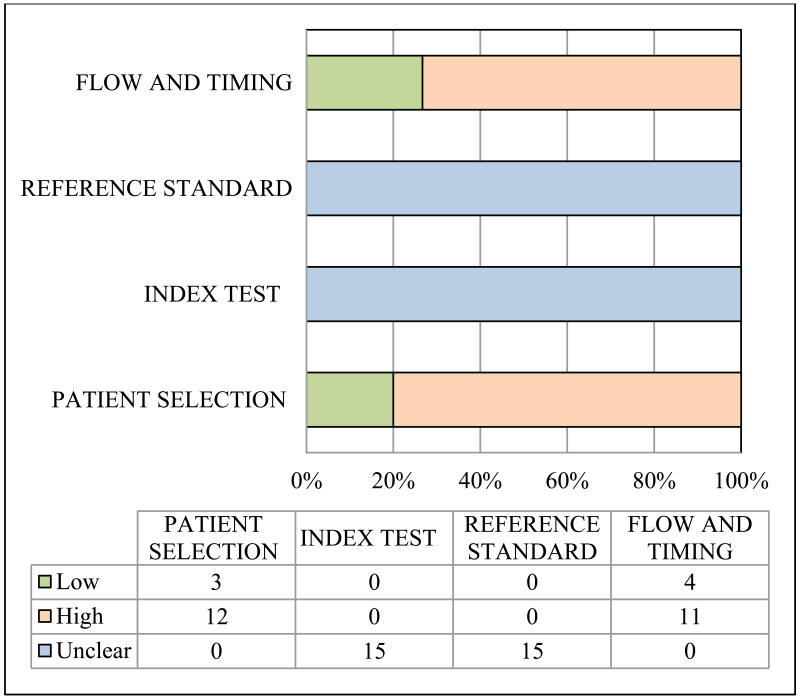
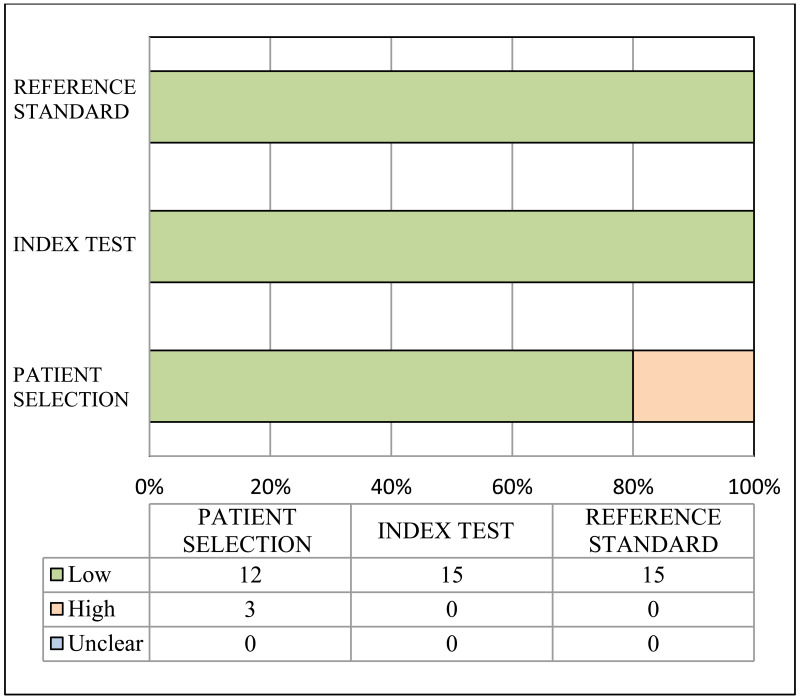
Risk of bias and applicability of population, index and reference tests to the main review questions were evaluated using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) (23). A list of the signalling questions used for QUADAS2 are presented in Appendix B.
3.7. Data extraction
The data were extracted from the selected studies by the two independent reviewers for each of the English (PB and KY) and Chinese articles (YL and TZ), using a pre-piloted data extraction form (Appendix C). In case of disagreement in the data extracted between the two reviewers, a deliberation that involved a third person (YS) was carried out.
During data extraction, articles from the same study sites with overlapping recruitment periods, enrolment criteria, and treatment types were considered as being part of one study. The lead reviewer for both English (PB) and Chinese (YL) articles then followed up with the corresponding author(s) from each of the article groups in order to understand if there was any patient overlap. If the authors explicitly stated in their article that there is overlap, or if the authors responded to the email inquiry confirming overlap, or if the author did not respond, then only the data extracted from the most recently published article was used in data analysis. If authors denied any patient overlap between articles then data extracted from all the articles within the group were used. In the case of a group of articles from the same study where some articles were published in Chinese and some in English, the latest English article was included in the data analysis sheet, unless a direct communication with the study authors directed the reviewers to use a different article in the group.
3.8. Data synthesis
All statistical analyses were performed using STATA 14.2 (Stata Corp LP, College Station, TX).
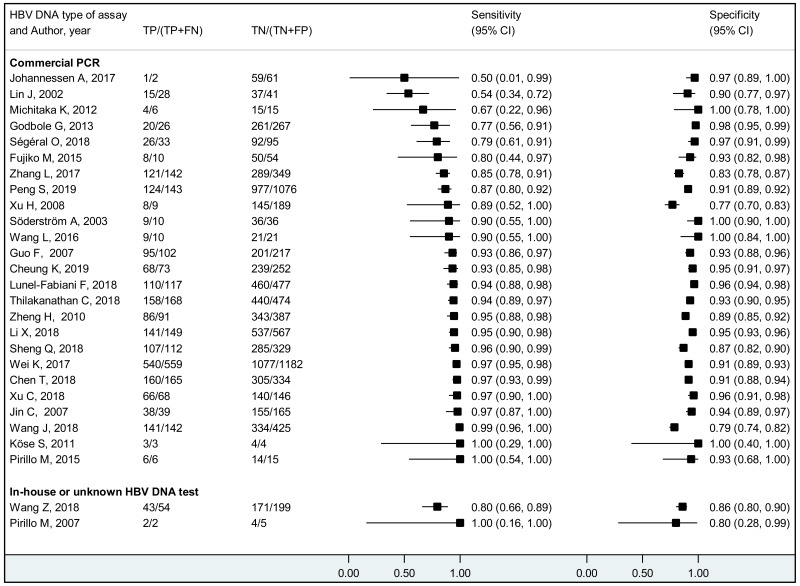
Sensitivity and specificity were estimated at each of different HBV DNA thresholds used in the included studies (≥20 000 IU/mL, ≥5 log10 IU/mL, ≥6 log10 IU/mL, ≥7 log10 IU/mL and ≥8 log10 IU/mL). In case a single study presented the results at multiple HBV DNA thresholds, all the different thresholds were used. In addition, for PICO2B and 2C, sensitivity and specificity were estimated specifically for each measure of prevention for HBV MTCT. The summary statistics were pooled only when there were at least three studies.
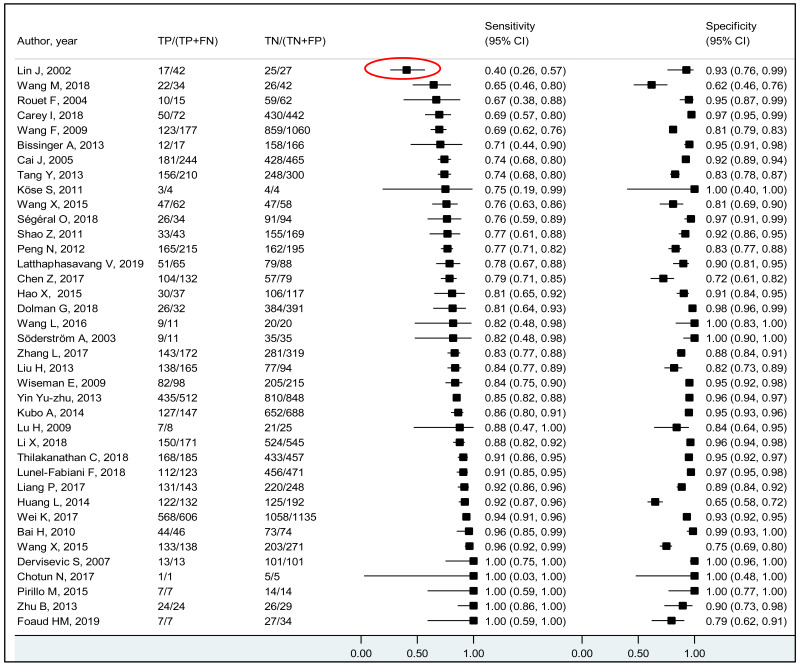
We performed bivariate analysis for studies allowing estimation of both sensitivity and specificity. Study-specific estimates of sensitivity and specificity along with their 95% CI were graphically presented in coupled forest plots. When there were at least three studies, the summary estimates for sensitivity and specificity along with their 95% CI were obtained using the DerSimonian–Laird bivariate random effects model. Positive and negative likelihood ratios (PLR, NLR) with 95% CIs were obtained from the pooled sensitivity and specificity. When there were <3 studies, the range of sensitivity, specificity, PLR, and NLR, were presented. Pre-test probabilities were estimated by pooling the proportion of pregnant women with CHB who had high viral loads. After the variance of the proportions was stabilized using Freeman–Tukey double arcsine transformation, these estimates were pooled using the DerSimonian–Laird random-effects model (24). Post-test probabilities were computed using the pre-test probabilities and the pooled PLR and NLR.
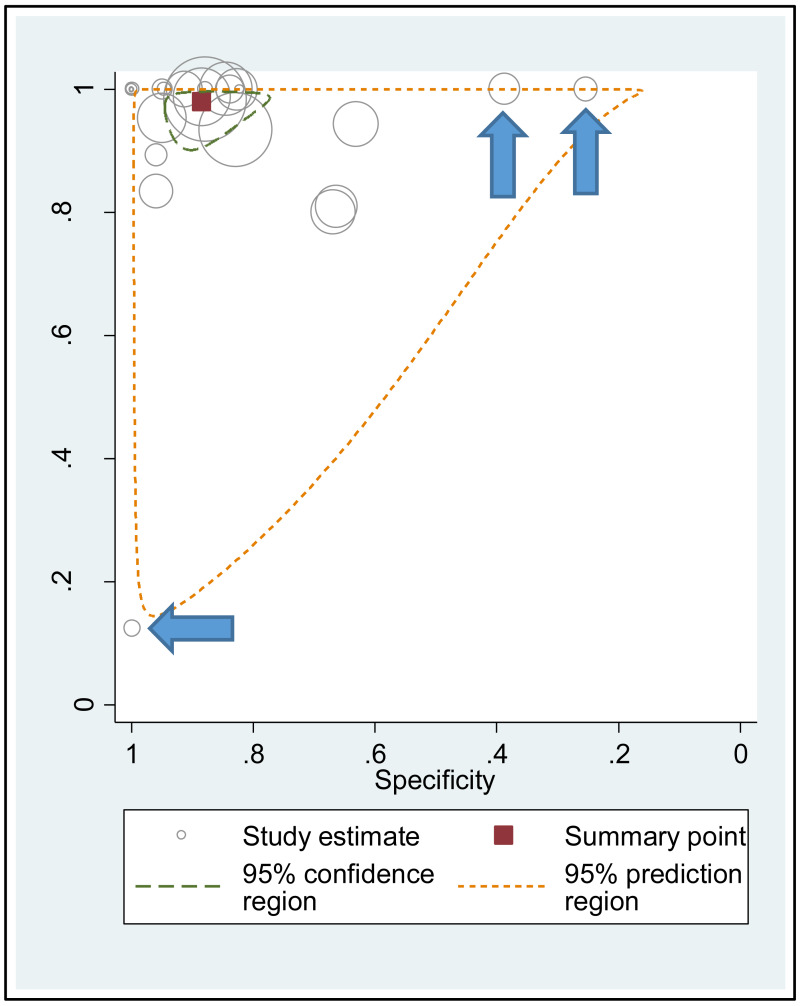
Heterogeneity in the estimates across the studies was visually assessed using: (i) coupled forest plots displaying study-specific estimates of sensitivity and specificity, and (ii) summary scatter plots. The summary scatter plots were presented without a summary receiver operating characteristic (SROC) curve for PICO2A and with a SROC curve for PICO2B and 2C. Characteristics of the outlier studies were narratively described. A sensitivity analysis was also performed after excluding these outlier studies.
Heterogeneity was also assessed statistically, by considering the following variables as a priori potential sources of heterogeneity: type of HBeAg assay, type of reference standard (commercial PCR vs other/not reported), mean/median maternal age, maternal virological characteristics (HIV/HCV/HDV coinfection status and HBV genotypes), WHO region, and study’s risk of bias (high vs low). In addition, for PICO2B and 2C, the measures of prevention for HBV MTCT were considered. Models fitted with and without the covariate were compared using likelihood ratio tests assuming equal variances. When there was good evidence (P<0.05) to support the heterogeneity, another model was fitted with separate variances and compared to the model with equal variances to understand if the heterogeneity observed could be due to the differences in variances between studies within a category rather than differences between categories of variables identified as potential sources of heterogeneity.
To integrate estimates from studies that provided data only for sensitivity or specificity, we performed univariate analyses using the DerSimonian–Laird univariate random effects model. When the study estimates could not be pooled (<3 studies), the range of sensitivity and specificity was presented.
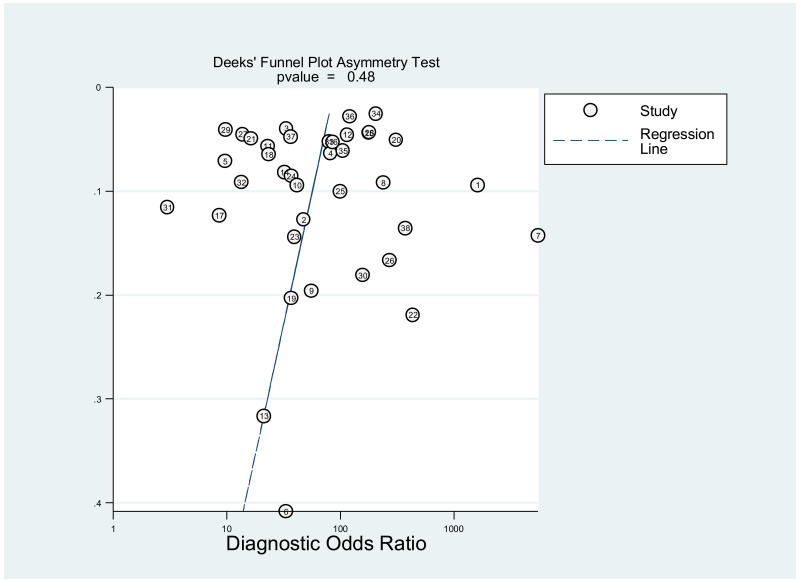
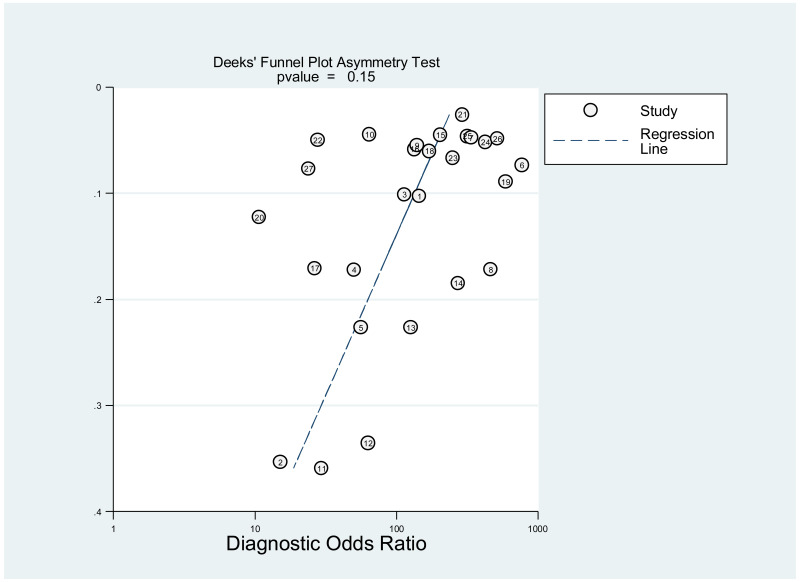
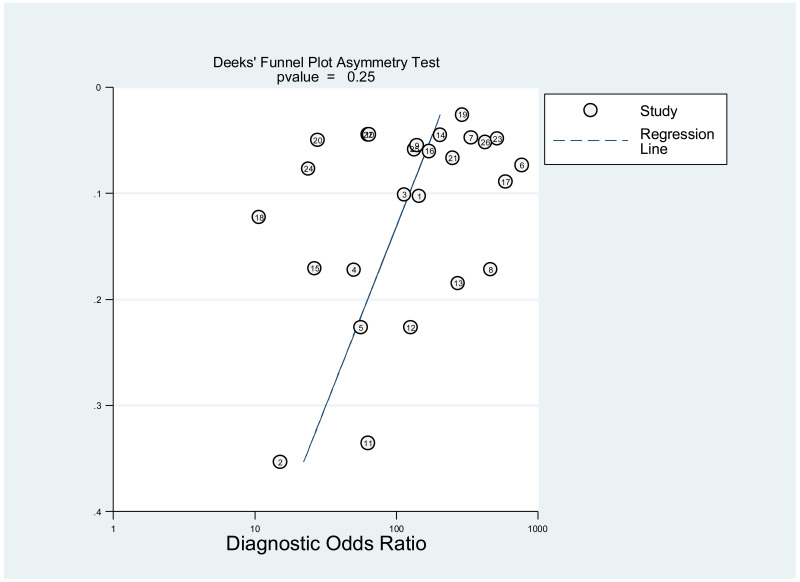
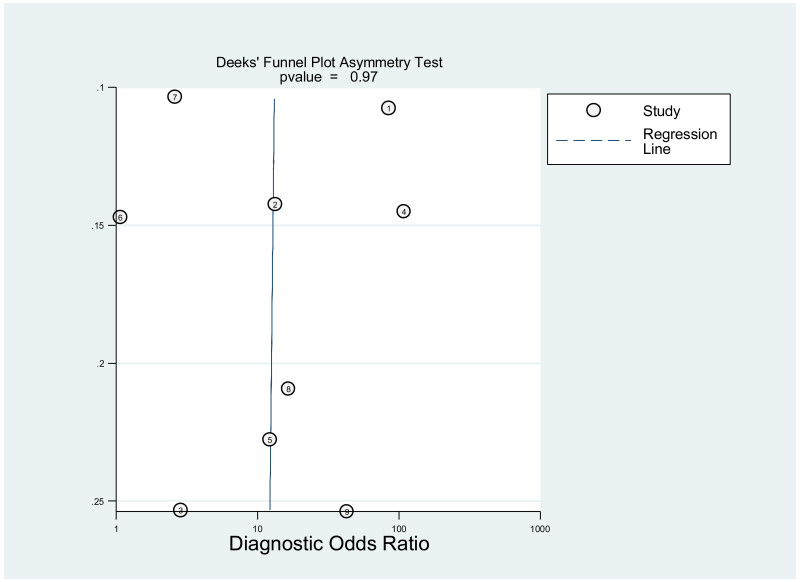
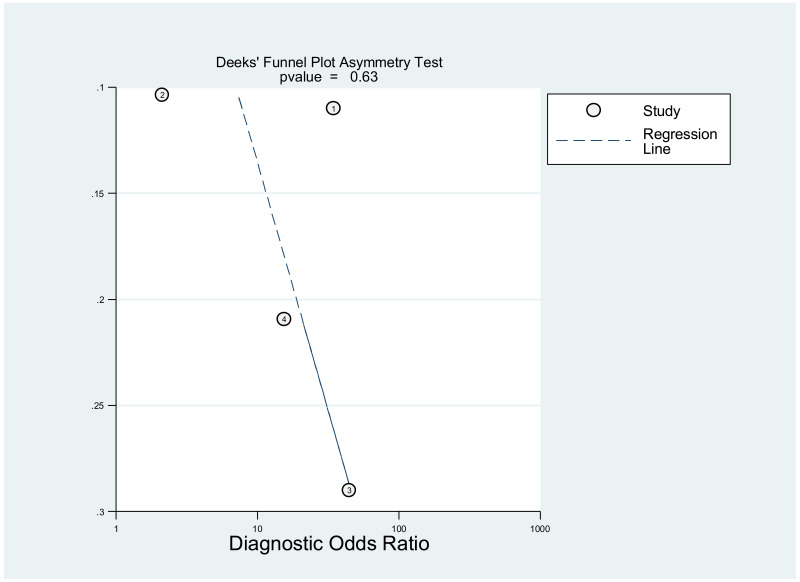
Publication bias was assessed using Deeks’ test, which was developed specifically for diagnostic accuracy reviews and is the method recommended in the Cochrane Handbook for Diagnostic Test Accuracy Reviews (25). It tests the asymmetry of the plot of log diagnostic odds ratio (lnDOR) against 1/effective sample size (ESS)1/2. The ESS is a function of the number of diseases (n1) and non-diseased participants (n2) ((4n1*n2)/(n1 + n2)) and this takes into account the numbers of diseased and non-diseased participants (26,27).
The post-hoc analysis 1 was conducted as below. For the studies using “IU/mL” as a unit for HBV DNA levels, MTCT risk was estimated for the following maternal HBV DNA levels during pregnancy: <4.00; 4.00–4.99; 5.00–5.99; 6.00–6.99; and ≥7.00 log10 IU/mL. For those using “copies/mL”, the maternal viral load was transformed into “IU/mL” (by dividing by the factor of 5) and risk was estimated for the following HBV DNA levels: <4.30; 4.30–5.29; 5.30–6.29; 6.30–7.29; and ≥7.30 log10 IU/mL. In case a single study presented the results at multiple HBV DNA thresholds, all different thresholds were used.
Once the HBV DNA level where the risk of MTCT was identified despite infants’ immunoprophylaxis, post-hoc analysis 2 was conducted. The sensitivity and specificity of the HBeAg test to diagnose this HBV DNA threshold were estimated using the method described above.
3.9. GRADE review process
For each examined PICO question, the quality of the evidence was evaluated using the Grading of Recommendations Assessment, Development and Evaluation methodology (GRADE) (28). We used this tool to evaluate: (i) the risk of bias; (ii) inconsistency (high heterogeneity); (iii) imprecision (confidence intervals); (iv) indirectness (use of surrogate outcomes); (v) reporting and publication bias; and (vi) other factors; for each of the outcomes. This eventually gave a score of high (further research is very unlikely to change the effect estimate), moderate, low or very low (all estimates are very uncertain). Decisions for the complex judgements within the GRADE table were made through study group consensus. The study group reviewers were supported in the process of completing this GRADE template through discussion and advice from a WHO-designated methodological expert, Professor Roger Chou (Oregon Health & Science University, USA). For this specific meta-analysis, the following rules were used to determine whether or not a group of studies had no serious, serious, or very serious issues with regard to GRADE criteria:
- -
GRADE scoring system:
As cohorts and cross-sectional studies can provide reliable evidence for diagnostic accuracy, strength of evidence was initially rated as high quality (29). Then, strength of evidence was lowered by one degree if there was “serious” and by two degrees if there was “very serious” risk of bias, inconsistency, indirectness or imprecision. The strength of evidence was similarly lowered by one degree if publication bias was “likely” and by two degrees if the bias was “very likely” (30).
- -
Risk of bias:
A study was considered as “high” overall risk of bias when multiple QUADAS-2 domains were rated as “high risk of bias”. Then, for each outcome, the number of studies with “high risk of bias” was counted. The risk of bias for the outcome was rated as “very serious”, “serious” or “not serious” when the proportion of studies rated as “high risk of bias” was >75%, >50–75% or ≤50%, respectively.
- -
Indirectness:
Indirectness is linked with the level of applicability of the study population, index test or reference standard to the review question. A study was considered as “high” overall concern about applicability when at least one out of the three QUADAS-2 domains was rated as “high concern about applicability”. Then, for each outcome, the number of studies with “high concern about applicability” was counted. Indirectness for the outcome was rated as “very serious”, “serious” or “not serious” when the proportion of studies rated as “high concern about applicability” was >75%, >50–75% or ≤50%, respectively (29).
- -
Imprecision:
Imprecision was considered “not serious” when an absolute range in the 95% confidence intervals (95% CI) for a pooled sensitivity or specificity was ≤20%. Imprecision was “serious” or “very serious” when the range was 20–40% or >40%. Moreover, when the cumulated sample size for all included studies was <30, it was categorized as “very serious”.
- -
Inconsistency:
Inconsistency was considered “not serious” when ≥75% of studies’ estimates were within +/–20% of the pooled estimate for an outcome. Inconsistency was considered “serious” or “very serious” when this proportion was 50–75% or <50%.
- -
Publication bias:
This was not assessed as part of the GRADE, because none of the studies, except one, was designed to assess diagnostic accuracy. Therefore, the analysis based on the diagnostic odds ratio seemed irrelevant.
5. Conclusion
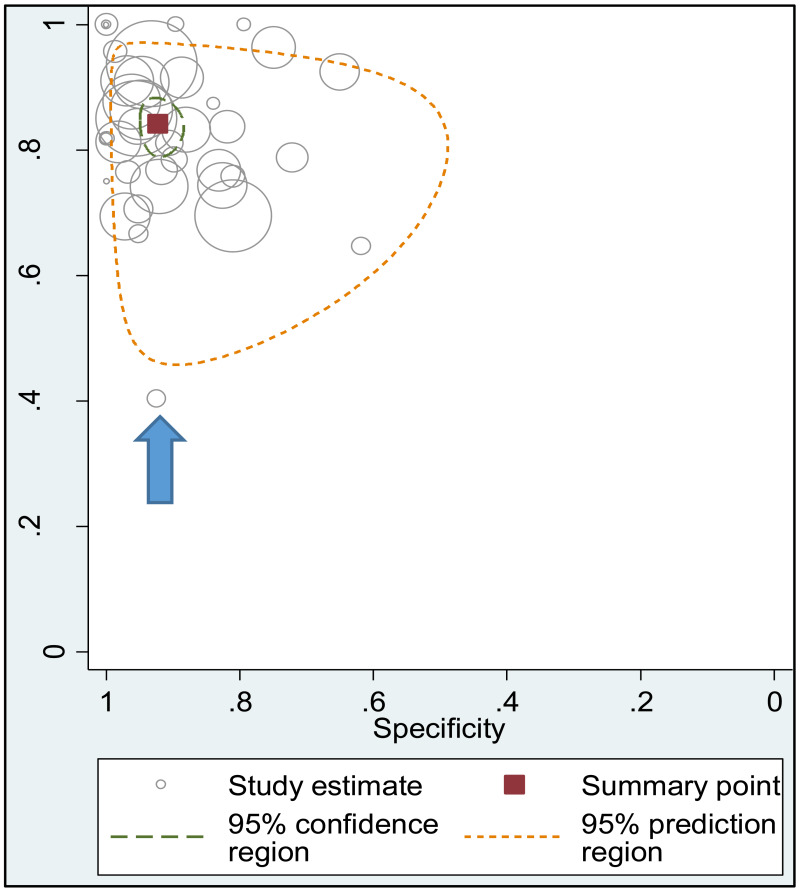
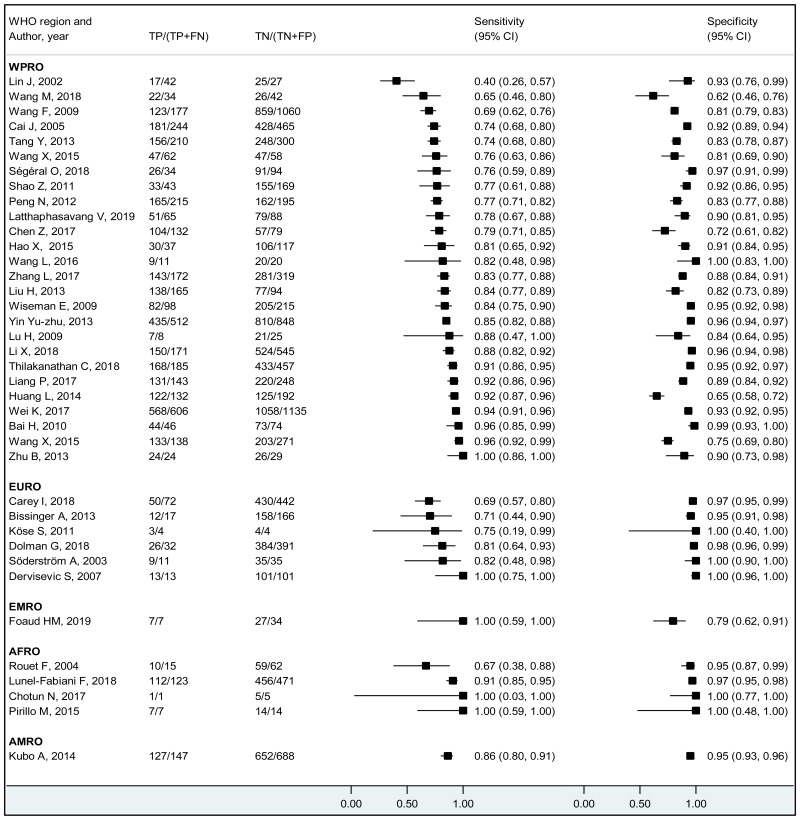
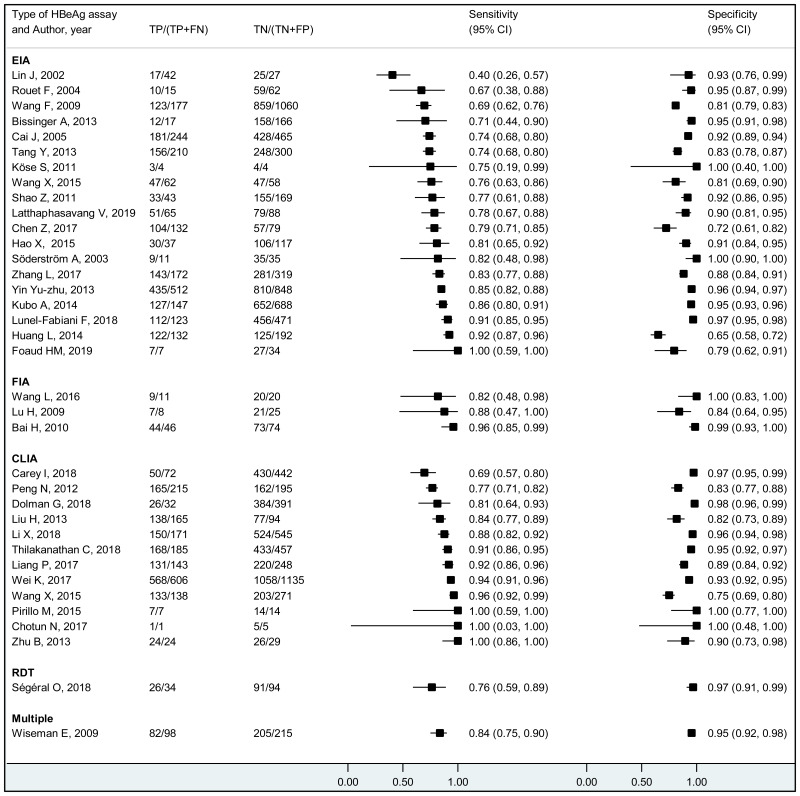
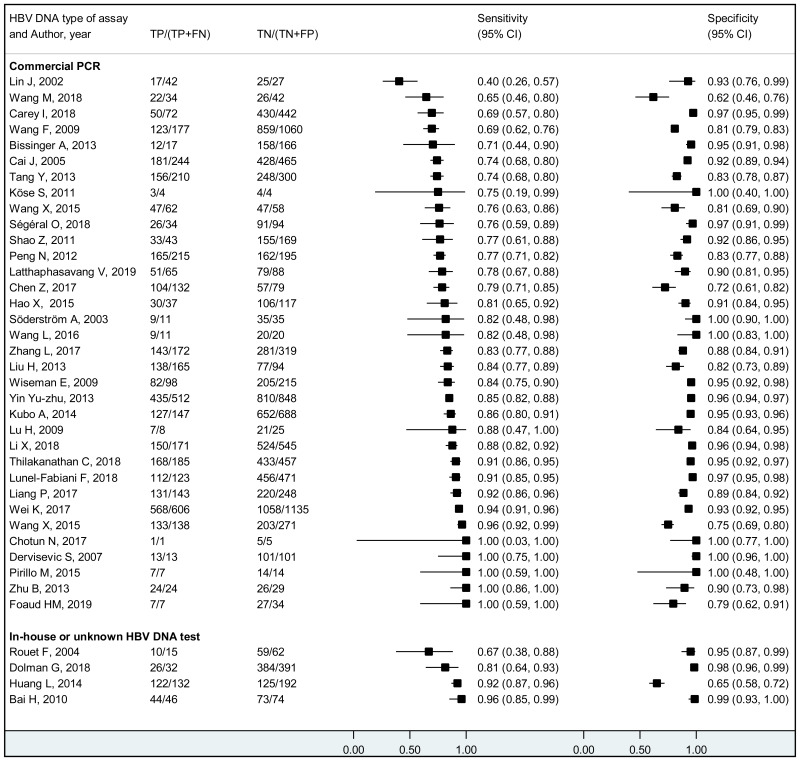
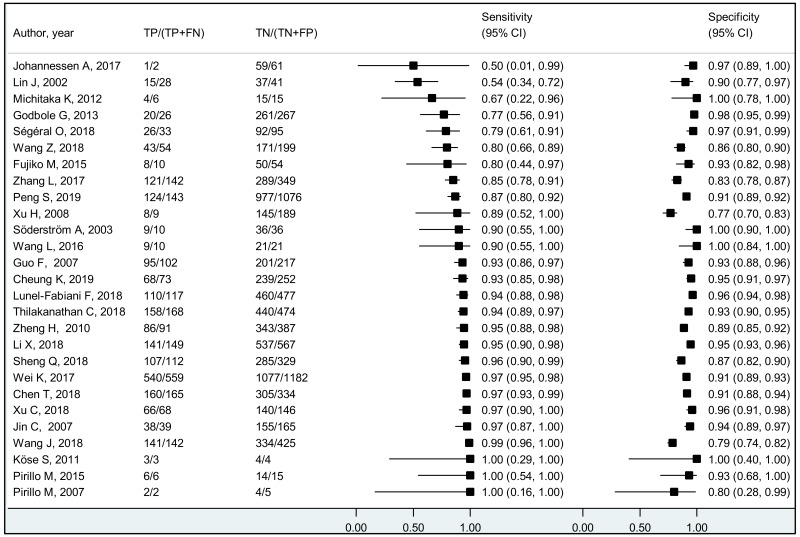
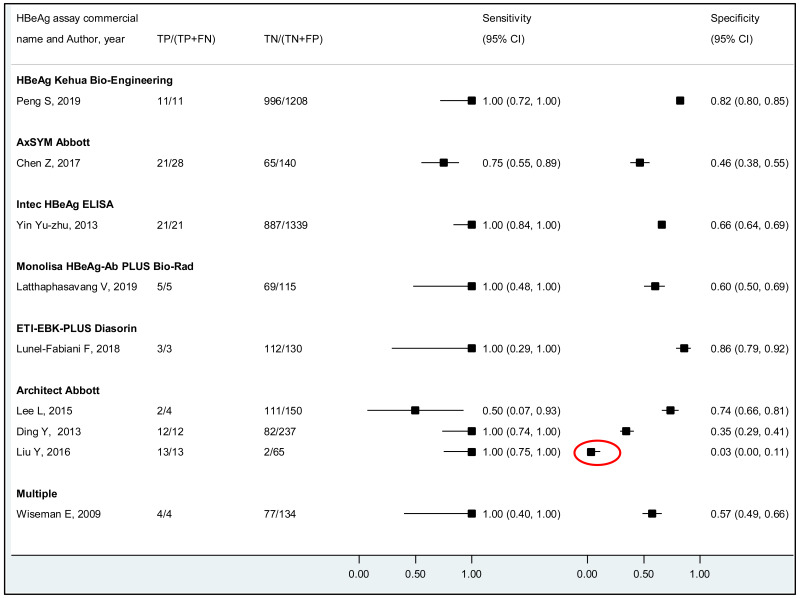
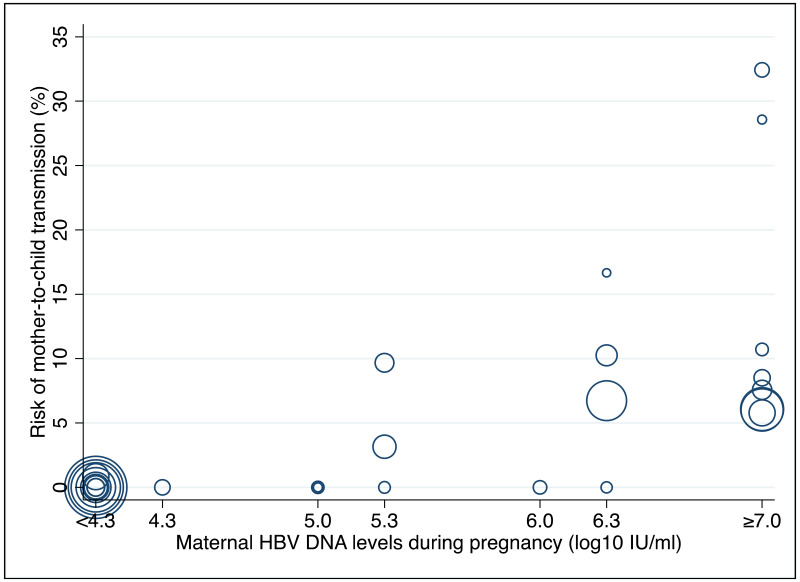
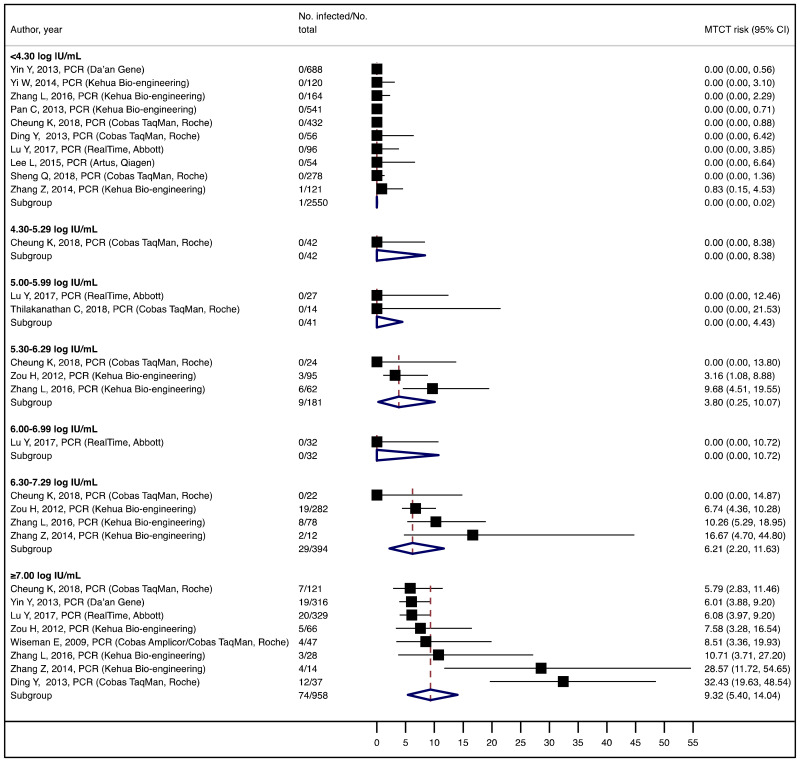
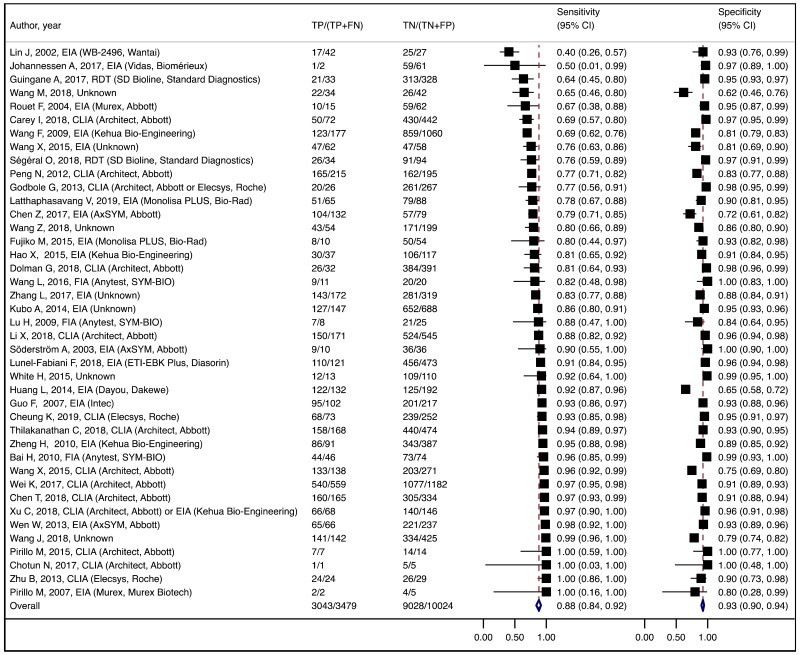
To our knowledge, this systematic review is the first to examine the diagnostic accuracy of the HBeAg test to diagnose high HBV DNA levels in pregnant women. Our results suggest that the risk of HBV MTCT, despite passive–active immunoprophylaxis, starts to increase at a maternal viral load of 5.3 log IU/mL (i.e. 200 000 IU/mL). The pooled sensitivity and specificity of HBeAg, obtained by the bivariate analyses, were: 84.2% (95% CI: 80.2–87.4%) and 92.3% (89.5–94.5%) to diagnose viral load of ≥5 log10 IU/mL; 92.0% (88.2–94.6%) and 92.7% (90.3–94.5%) for ≥6 log10 IU/mL; and 98.0% (93.3–99.4%) and 88.5% (80.7–93.4%) for ≥7 log10 IU/mL, respectively. We evaluated the performance of HBeAg for three other different HBV DNA thresholds (≥20 000, ≥200 000 and ≥8 log10 IU/mL). Irrespective of these different HBV DNA cut-off levels, pooled sensitivity and specificity were constantly higher than 80%, with the lower boundary of the 95% CI exceeding 75%. The univariate analyses provided similar results, supporting the robustness of these estimates. As expected, the sensitivity improved with increasing HBV DNA threshold whereas the specificity decreased.
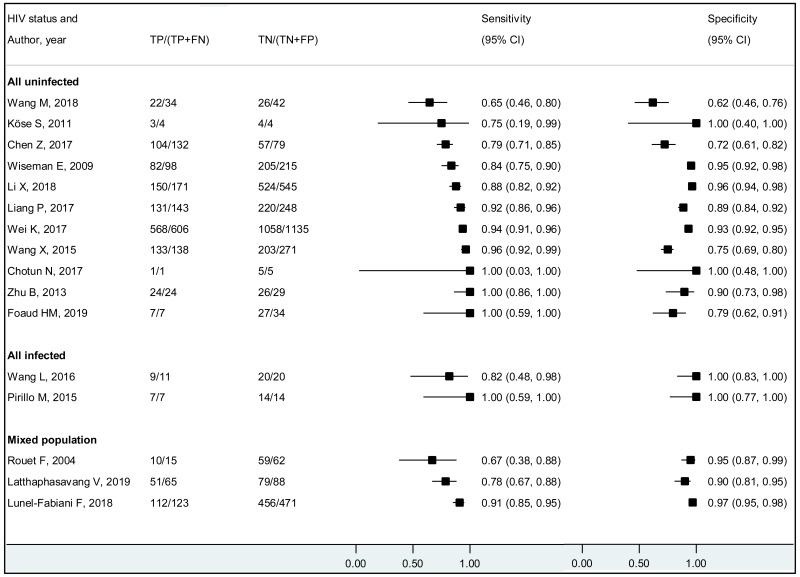
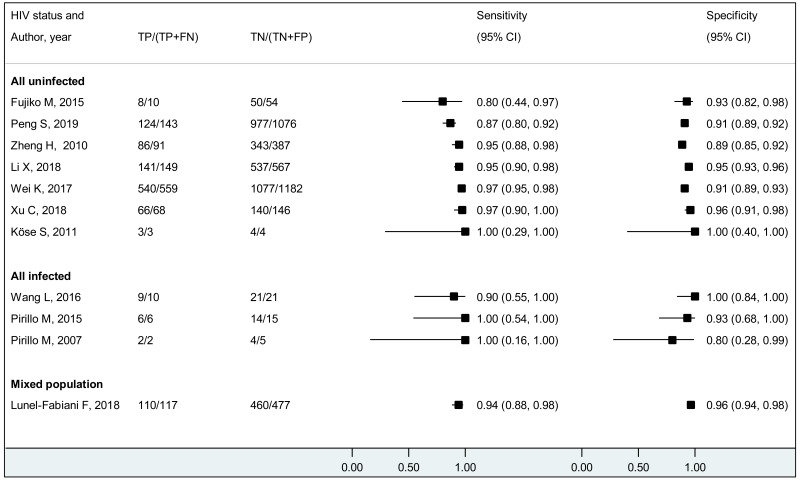
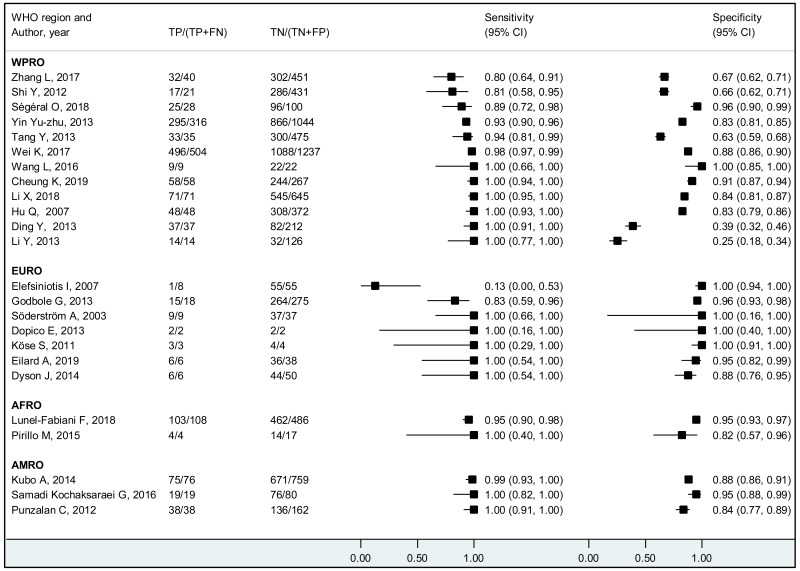
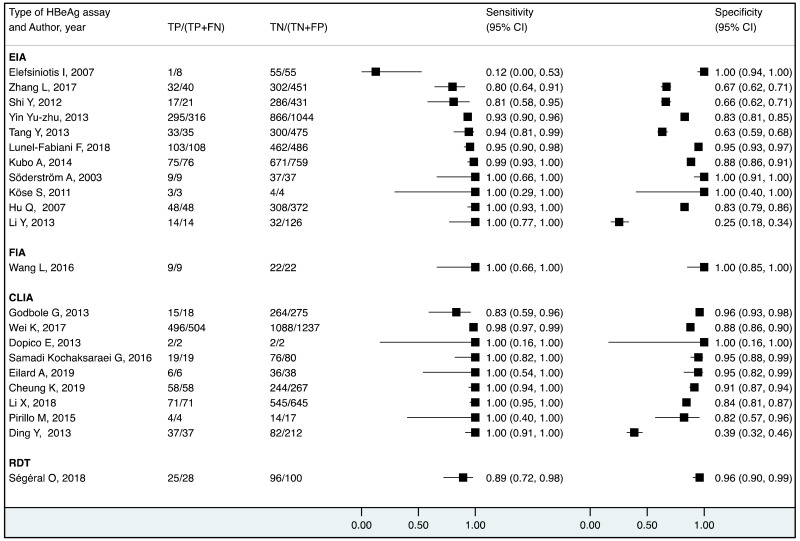
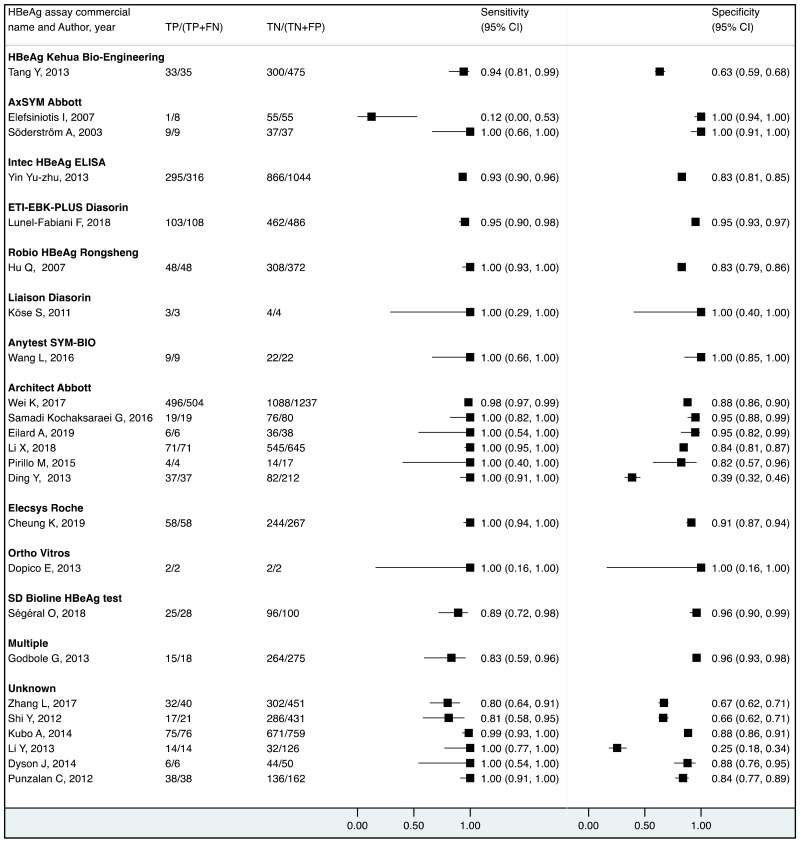
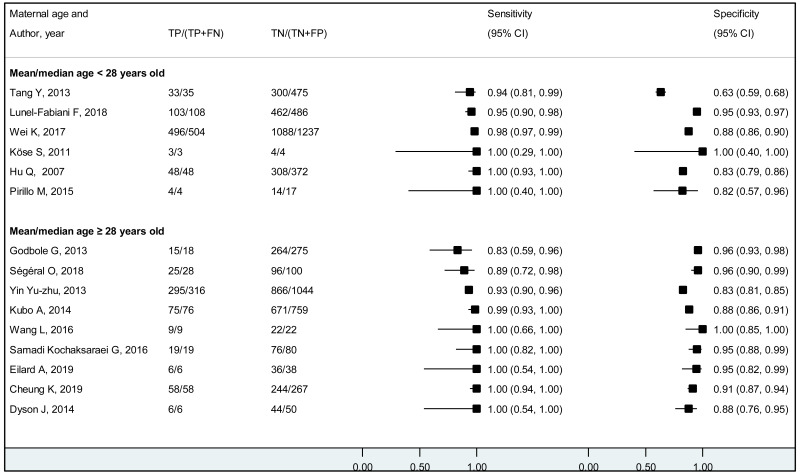
We found evidence that the performance of HBeAg during pregnancy to identify pregnant women with a high viral load differed according to the WHO region, type of HBeAg and maternal age. However, there was no evidence that its performance varied according to HBV DNA NAT or maternal HIV status.
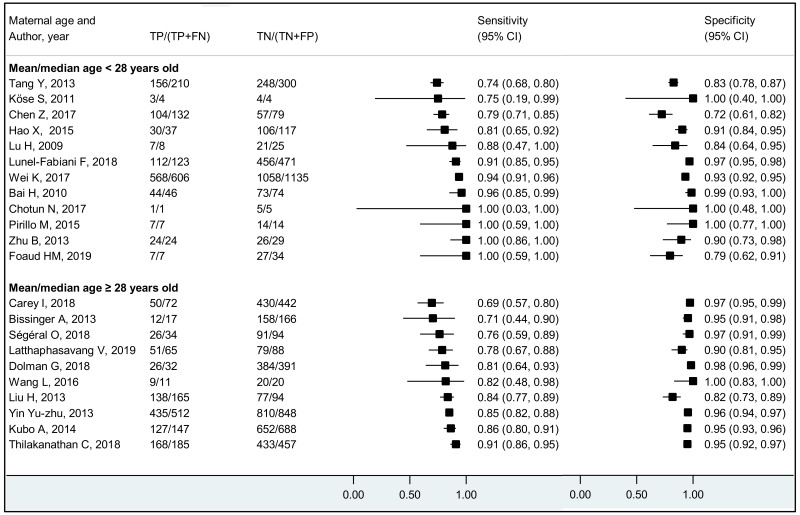
The performance of HBeAg differed significantly between lower (<28 years) and higher (≥28 years) mean/median maternal age reported in each study; younger maternal age was associated with higher sensitivity and lower specificity to diagnose HBV DNA levels of ≥5 and ≥6 log10 IU/mL, compared to higher maternal age (≥28 years). Although the difference was not significant, a similar tendency was observed for the cut-off of ≥7 log10 IU/mL. Since spontaneous loss of HBeAg occurs over time, the prevalence of HBeAg is higher in younger HBsAg-positive women than in older HBsAg-positive women (120), and this may explain the difference in performance of HBeAg according to maternal age. The natural history of CHB varies according to geographical area, particularly between Asia and sub-Saharan Africa, which both carry a high HBV-related disease burden. Historically in both areas, the majority of CHB occurs during childhood either perinatally from infectious mothers or horizontally from household members. In Asia, a substantial proportion (about 40%) of children who are chronically infected with HBV continue to carry HBeAg and high viral load beyond their adolescence (121) while in Africa, spontaneous HBeAg loss often occurs at younger age and only 10–20% of HBV-infected women of childbearing age carry HBeAg (122). This difference might be due to varying frequency of emerging basal core promoter (BCP) or precore (PC) variants, which abolish or reduce HBeAg production without affecting the capacity of HBV to replicate (123). We found a similar high sensitivity of HBeAg to diagnose high viraemia in both regions: 83.9% (79.2–87.7%) in Asia and 88.0% (70.9–95.7%) in Africa for HBV DNA levels ≥5 log10 IU/mL, respectively; 92.6% (88.4–95.3%) and 93.7% (87.9–96.8%) for HBV DNA levels ≥6 log10 IU/mL, respectively. However, the specificity of HBeAg to diagnose high maternal viral loads tended to be lower in Asia (Western Pacific) compared with Africa: 89.3% (85.6–92.1%) and 96.6% (94.3–98.0%) for HBV DNA levels ≥5 log10 IU/mL; 91.1% (88.4–93.2%) and 96.2% (94.3–97.5%) for HBV DNA levels ≥6 log10 IU/mL, respectively.
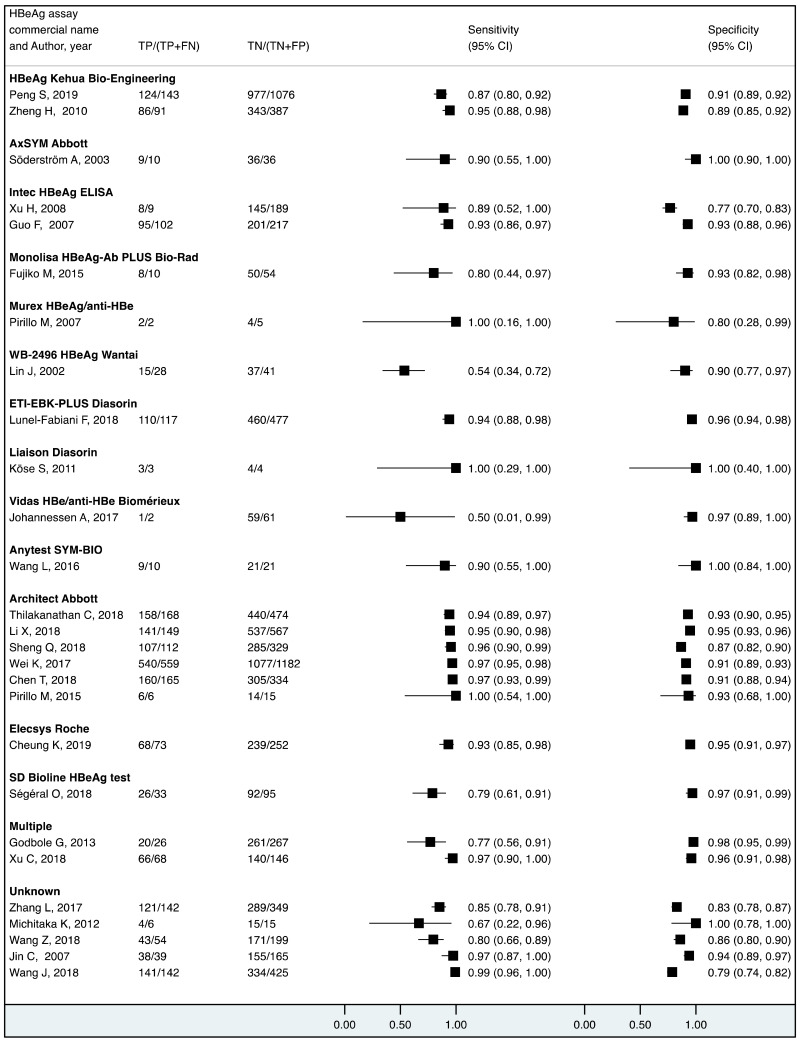
More recent types of HBeAg assays such as CLIA tended to have a higher sensitivity for diagnosing viraemia than EIA: 89.1% (95% CI: 83.2–93.0%) and 79.4% (74.0–84.0%) to diagnose viral load ≥5 log10 IU/mL; and 94.4% (81.3–98.5%) and 98.8% (93.7–99.8%) to diagnose viraemia ≥7 log10 IU/ml, respectively. This might be related to improved analytical sensitivity of CLIA, in comparison with EIA, to detect HBeAg. Although there was only one study that evaluated RDT, this had lower clinical sensitivity compared to the laboratory-based immunoassays (76.5% and 89.3% to diagnose high HBV DNA levels of ≥5 log10 and ≥7 log10 IU/mL, respectively). Low clinical sensitivity of commercially available HBeAg RDT was also found to be related to its low analytical sensitivity compared to CLIA (17).
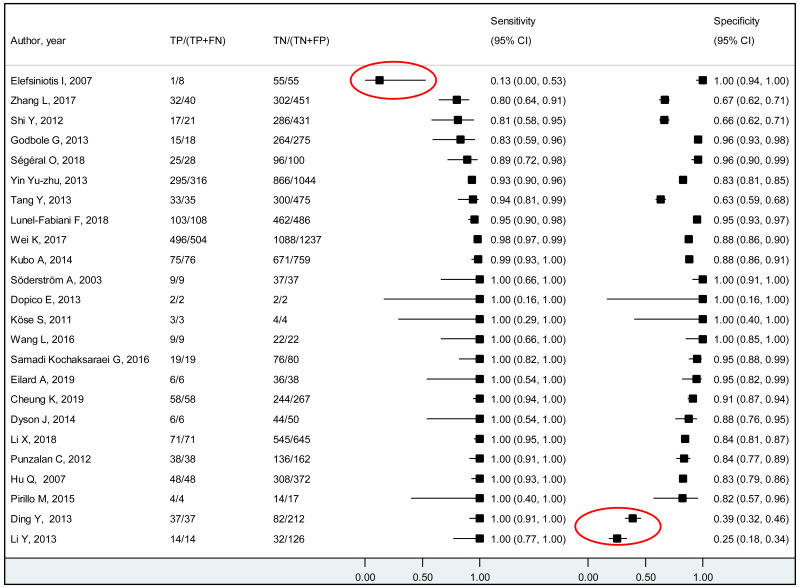
A few outlying studies were identified: one study for ≥5 log10 IU/mL and three studies for ≥7 log 10 IU/mL. Compared to other studies, these outliers tended to have either “lower sensitivity and higher specificity” (50,70), or “lower specificity with higher sensitivity” (45,68). Although we did not perform further assessment, this might be related to multiple factors such as low HBeAg prevalence (1.6%) in HBsAg-positive pregnant women in a Greek study (50) or the difference in analytical sensitivity of the HBeAg tests used in these outlier studies (68,70). The sensitivity analyses excluding these outliers did not alter the interpretation of these results.
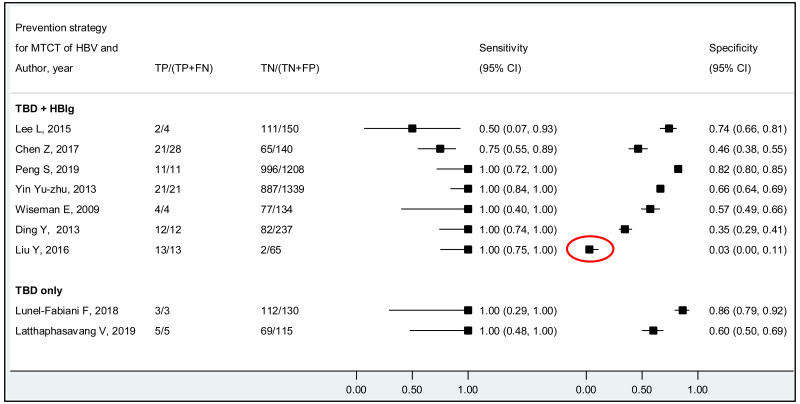
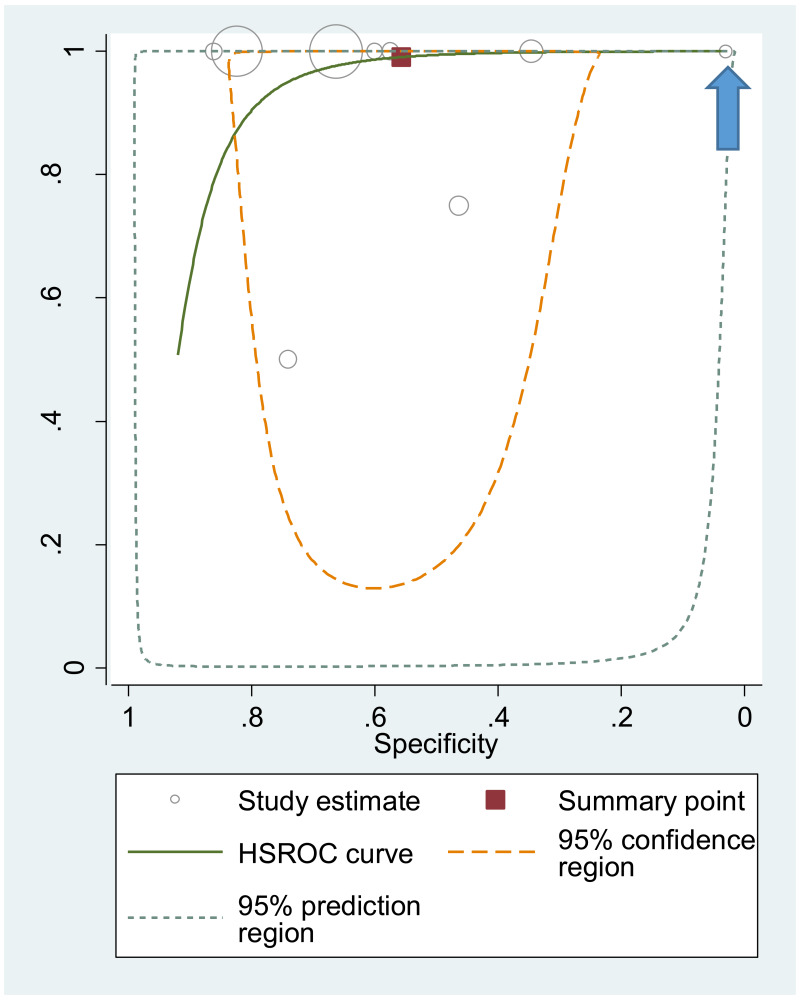
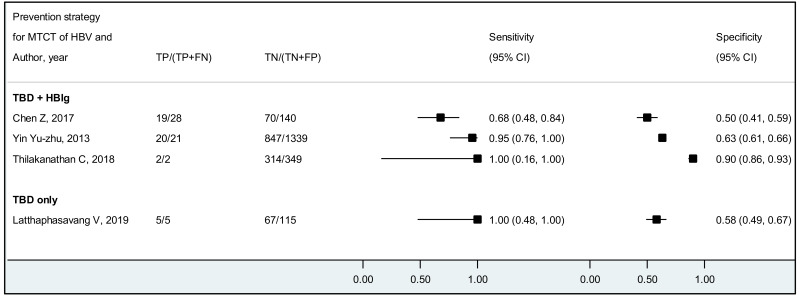
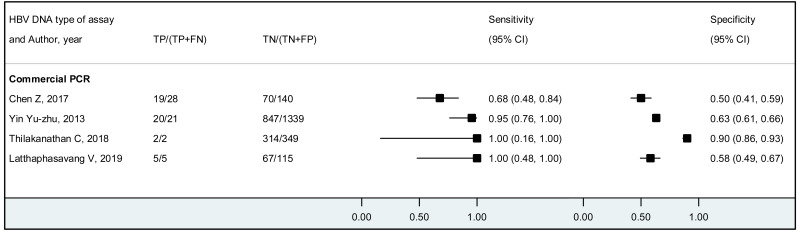
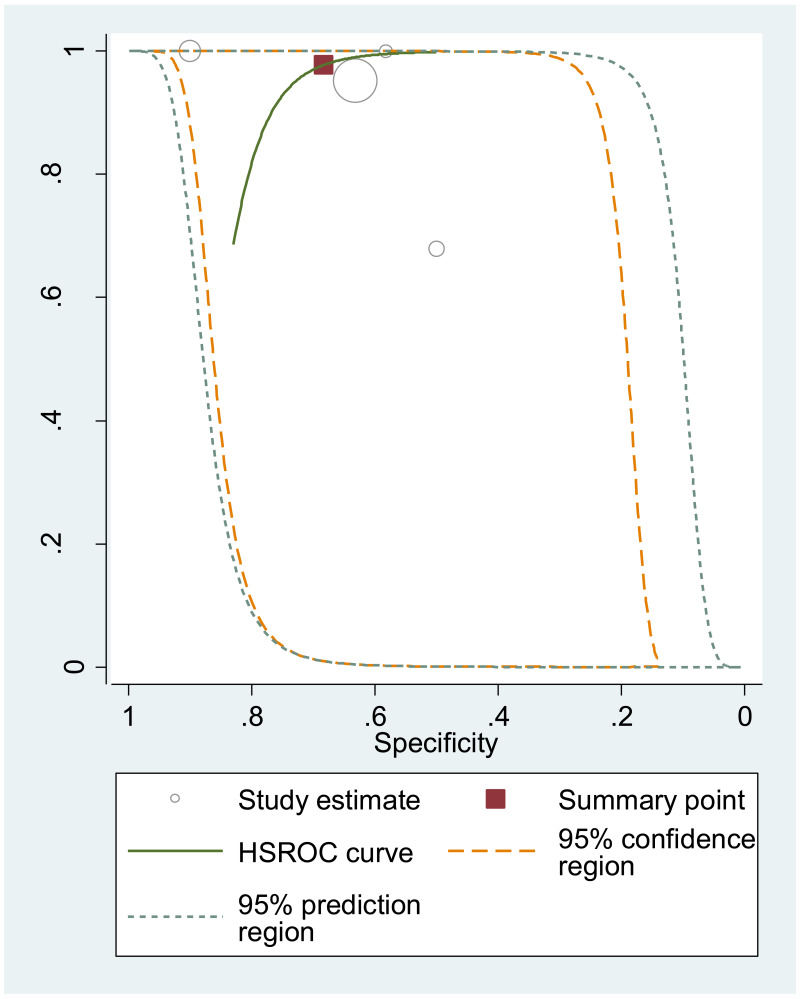
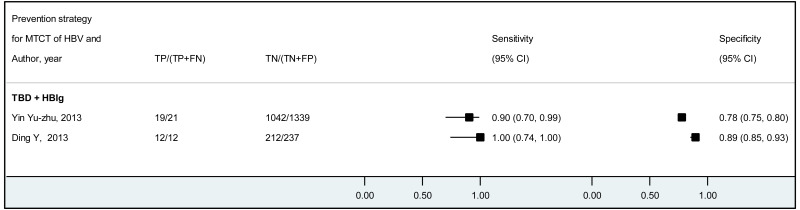
The systematic review addressed two additional questions: the performance of HBeAg detection and HBV DNA quantification during pregnancy to predict an MTCT event, defined as HBsAg positivity in infants aged 6–12 months. These analyses were stratified by the type of preventive measures provided to mother–child pairs. We found that the pooled sensitivity and specificity of maternal HBeAg during pregnancy to predict MTCT despite infant immunoprophylaxis were 99.1% (95% CI: 61.8–100%) and 55.7% (34.0–75.5%). The pooled sensitivity and specificity of maternal HBV DNA levels ≥5 log IU/mL was 97.7% (95% CI: 42.9–100.0%) and 68.4% (95% CI: 48.6–83.2%); and the sensitivity and specificity range of maternal HBV DNA levels ≥7 log IU/mL were 90.5% to 100.0% and 77.8% to 89.5%, respectively. Although no formal assessment was performed, these results might indicate that: (i) both HBeAg and high HBV DNA levels, measured during pregnancy, have high sensitivity to predict the risk of immunoprophylaxis failure; and (ii) the specificity is low for HBeAg and moderate for high HBV DNA levels.
The strength of the evidence for the performance of HBeAg to identify pregnant women with viral loads ≥5, ≥6, and ≥7 log 10 IU/mL were high. Although most studies had excluded women coinfected with either HIV, HCV or HDV, this did not constitute a serious risk of bias. There was no evidence of publication bias; however, only one study was designed to evaluate diagnostic accuracy, which might have made the conventional assessment of publication bias, through assessing small sample effects, less meaningful. Moreover, since essential information for diagnostic studies (e.g. blinding) were missing in the majority of studies, the assessment of the quality of studies was difficult for the index test and reference standard sections of the QUADAS2.
For the performance of HBeAg to predict MTCT, the strength of evidence was low due to high risk of bias in most studies and serious imprecision of the pooled estimates, probably due to the small number of included studies. When stratified by prevention of MTCT regimen, evidence was low for the timely birth dose plus HBIG regimen and could not be graded for timely birth dose only regimen (without HBIG) because there was not enough data available.
The strength of evidence for the overall performance of maternal HBV DNA levels ≥5 log 10 IU/mL was low due to inconsistency in the estimates and serious imprecision in the pooled estimates. When stratifying by prevention of MTCT strategy, evidence was low for timely birth dose plus HBIG for the same reasons as stated above, and there was not enough data to grade the evidence for the timely birth dose only strategy. For maternal HBV DNA levels ≥7 log 10 IU/mL, all included studies reported using the timely birth dose plus HBIG strategy and there was not enough data to grade evidence for specificity. Concerning sensitivity, evidence was low because of very serious imprecision in the pooled estimate, which may be due to the low number of studies included to answer this objective.
As a strength of this study, the literature was systematically searched through both English- and Chinese-language databases, and independently reviewed by two investigators for each language (a total of four reviewers). Duplicate publications were carefully checked and excluded from the analysis to avoid biased estimates.
As a limitation, this systematic review was primarily designed to accomplish the primary objective (PICO2A); we thus included only studies that measured both HBeAg and HBV DNA during pregnancy to answer the secondary objectives. Of the studies eligible for the PICO2A, we further selected those that followed infants to ascertain MTCT end-points to answer two additional questions (PICO2B and PICO2C). Consequently, we might have missed a few of the eligible studies for PICO2B (e.g. a study evaluating maternal HBeAg during pregnancy and infant HBsAg at 6 months without doing maternal HBV DNA) or PICO2C (e.g. a study evaluating maternal HBV DNA and infant MTCT end-point, without assessing HBeAg during pregnancy). Because of these limitations, care must be taken when interpreting PICO2B and PICO2C.
5.1. Implications for practice
This study suggests that risk of HBV MTCT, despite passive–active immunoprophylaxis, starts to increase at a maternal viral load of 5.3 log IU/mL. This threshold should be theoretically lower when the PMTCT strategy does not include HBIG (e.g. timely birth dose alone).
With the high strength of the evidence observed, this study suggests that HBeAg might be a good alternative marker to HBV DNA NAT to diagnose high HBV viral load during pregnancy. Moreover, although the strength of the evidence was low, the systematic review found high sensitivity (99.1% (95% CI: 61.8–100%)) of HBeAg during pregnancy to predict immunoprophylaxis failure in infants (MTCT despite administration of birth dose vaccine and HBIG), with a poor specificity of around 55%.
The findings are particularly relevant in countries where there is limited access to HBV DNA NAT. Even though the HBeAg test may perform less well than HBV DNA NAT to identify pregnant women with an elevated risk of MTCT, other parameters (lower costs, improved access to testng and uptake, better linkage to care, and greater feasibility) may favour its use in certain contexts.
5.2. Implications for research
The vast majority of the included studies were from the Western Pacific Region (WPR: 65.9%), followed by the European Region (EUR: 18.3%), African Region (AFR: 7.3%), the Americas (AMR: 6.1%), and only one study each from South-East Asia (SEAR: 1.2%) and Eastern Mediterranean Region (EMR: 1.2%). We need additional research, particularly outside East Asia. There was no study that included only HCV- or HDV-coinfected women. Only a few studies provided the estimates for HIV-coinfected mothers; we did not find any difference in performance of HBeAg to diagnose high viraemia according to HIV status. As there were only a few studies that assessed viral genotype, we could not investigate the performance of HBeAg during pregnancy in different HBV genotypes.
The use of RDT is more attractive than laboratory-based immunoassays, because the former may be less expensive, faster, easier to perform, and thus more feasible than the latter in a peripheral laboratory in resource-limited contexts. We identified only one study evaluating the performance of RDT during pregnancy, and its sensitivity tended to be lower than that of EIA or CLIA. We need additional studies to evaluate the performance of RDT in pregnant women; but we may also need improvement of analytical sensitivity of RDT to detect HBeAg. The development and evaluation of other low-cost molecular assays or serological markers (e.g. hepatitis B core-related antigen (HBcrAg)) is also highly warranted (124).