FAQs
Questions and answers for common NCBI Datasets questions
FAQs
For information about the deprecation and retirement of the NCBI Datasets command-line tools v13.x and older, and API v1, see the CLI v13.x (API v1) FAQs.
How do I download SRA data?
Unfortunately, retrieval of SRA data, including retrieval of SRA data by BioProject accession, is not supported by NCBI Datasets.
To download SRA data for a given BioProject, we recommend using the SRA Toolkit. For example, to obtain SRA data in FASTQ format for the BioProject accession PRJNA648656, try the following:
- Download and install the SRA Toolkit.
- Run
prefetch PRJNA648656to download SRA data in .sra format - Run
fasterq-dump PRJNA648656to extract FASTQ files from the downloaded .sra files.
For more information, see the SRA Tools Wiki pages, Downloading SRA Toolkit and How to use prefetch and fastwerq-dump to extract FASTQ-files
Where is the data I requested?
Your data is in the subdirectory ncbi_dataset/data/ within the zip archive you downloaded.
I still can’t find my data, can you help?
We have identified a bug affecting Mac Safari users. When downloading data from the NCBI Datasets web interface, you may see only a README file after the download has completed (while other files appear to be missing). As a workaround to prevent this issue from recurring, we recommend disabling automatic zip archive extraction in Safari until Apple releases a bug fix. For more information, visit: Mac Safari zip archive bug
What file formats can be downloaded using NCBI Datasets?
Datasets offers the following file formats (if available for the requested query):
- Sequence files in FASTA format: genomic/gene, transcript and protein nucleotide sequences
- Annotation files: GTF, GFF3, and GBFF
- Metadata files: JSON and JSON Lines
What is a data package?
A “data package” is an NCBI Datasets zip archive that contains sequence, annotation, metadata and other biological data. For more information, see Data packages.
What is a dehydrated data package and what is rehydration?
A dehydrated data package is a zip archive that contains only metadata and the location of sequence and other data files on NCBI servers.Rehydration is the process of downloading the data itself.
Downloading a dehydrated data package and rehydrating it is the best way to download large genome data packages containing either > 1,000 genomes or > 15 GB of data. For more information, see our How-to guide on how to download large genome data packages.
What input date formats are supported by the command-line tools?
The NCBI Datasets CLI supports many input date formats by using datepase.ParseAny
to parse input date strings. However, we recommend using ISO 8601 standard YYYY-MM-DD to avoid any doubt that the date you provide is misinterpreted.
Why do gene counts differ when comparing taxonomy and species pages to the gene table?
Gene counts on the taxonomy and species pages are derived from the annotation report. The annotation report and other genome annotation files represent a snapshot of the genome at the time of genome annotation. In contrast, the gene table and gene data obtained from the datasets (datasets download gene...) contains current gene data, including unannotated genes, genes created after the last annotation, as well as any updates made to existing genes after the last annotation. For some model organisms, particularly human, frequent manual curation means that current gene data is likely to differ compared to the most recent annotation.
What are atypical genomes?

Atypical genomes are genomes with one or more problems that have been identified by NCBI relating to quality, unusual size, or other flaws in the genome assembly. See atypical assemblies for the list of problems that result in an affected genome being designated as atypical.
On individual genome pages, atypical genomes can be identified by the presence of a warning icon, consisting of a yellow triangle containing an exclamation point, and the type of genome problem at the top of the page (see image above).
On genome table pages, atypical genomes with one or more problems are identified with a warning icon, consisting of a yellow triangle containing an exclamation point, next to the genome assembly name. The type of genome problem(s) is shown when you hover on the warning icon. Atypical genomes can be excluded from the genome table display by selecting the “Exclude atypical genomes” checkbox in the Filters section of the page.
The assembly data report includes two fields that indicate whether a genome is atypical and the specific type of genome problem. When a genome problem has been identified, atypical.is_atypical is true, and atypical.warnings will include the type of genome problem(s).
How does NCBI decide which genomes to annotate?
Only genomes with assemblies that are publicly available in INSDC (DDBJ, ENA or GenBank ) are considered for inclusion in RefSeq and processing by the eukaryotic genome annotation pipeline. NCBI makes this selection based on several factors. For more information, see Genomes Selected for RefSeq Annotation.
How do I download plasmid sequences?
NCBI Datasets currently only provides access to plasmids that are part of a genome assembly. The comprehensive set of RefSeq plasmid sequences, including plasmids that are not part of a genome assembly, are available from the genomes FTP site at this path: https://ftp.ncbi.nlm.nih.gov/genomes/refseq/plasmid/
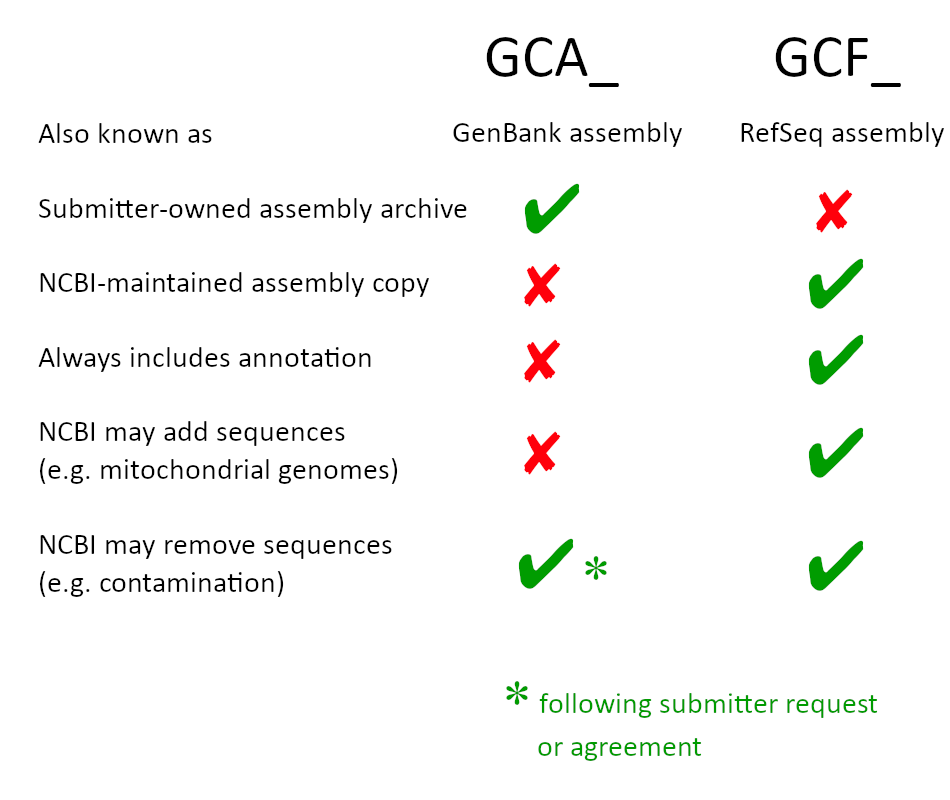
What is the difference between a GenBank (GCA) and RefSeq (GCF) genome assembly?
A GenBank (GCA) genome assembly contains assembled genome sequences submitted by investigators or sequencing centers to GenBank or another member of the International Nucleotide Sequence Database Collaboration (INSDC). The GenBank (GCA) assembly is an archival record that is owned by the submitter and may or may not include annotation. A RefSeq (GCF) genome assembly represents an NCBI-derived copy of a submitted GenBank (GCA) assembly. RefSeq (GCF) assembly records are maintained by NCBI. In some cases the RefSeq (GCF) assembly may not be completely identical to the GenBank (GCA) assembly due to assembly improvements made by NCBI staff. All RefSeq (GCF) genome assemblies include annotation.
What is changing with the NCBI Datasets API rate limits in January 2025, and how will it affect programmatic access, including the command-line tool?
Starting in January 2025, NCBI Datasets will implement a new rate limit for all programmatic access, including the NCBI Datasets command-line tools. There is no action needed from users that are accessing NCBI Datasets through the web. The rate limit will be set at 5 requests per second (rps) by default. This change is being made to ensure that all users, including those utilizing the command-line tool, can access the service smoothly and without interruptions.
Can I make more than 5 requests per second?
Yes, you can exceed the default rate limit of 5 requests per second (rps) using an NCBI API key. With an API key, you can make requests at a rate of 10 rps.If I already have an NCBI API key, do I need to request a new one to work with NCBI Datasets?
No, if you already have an NCBI API key, you can continue using the same API key with NCBI Datasets.
How do API keys benefit both users and NCBI?
API keys allow users to increase their request rate limit and serve as a valuable communication tool between users and NCBI. They enable NCBI to monitor and troubleshoot problems more effectively, ensuring a smoother experience for users. In case of issues or questions, having an API key associated with your requests can aid in quicker issue resolution and better user support. It also helps NCBI maintain the quality and reliability of its services.
