Data We Expect
Table of Contents
Introduction
Structural Variation (SV) can be complex to represent. Current technologies rarely provide base pair resolution for variant breakpoints. The experimental methods used determine the extent of this uncertainty. This in turn affects the way you report variants. Data representation can largely be broken down into the following categories:
- Variants for which we can define a minimal region that is definitely affected, but are unable to define precise breakpoints - only a range of coordinates within which the breakpoints likely occur (e.g., array CGH, SNP array)
- Variants for which a defined region of the genome is known to contain the variant, but the exact location of the variant and the breakpoints within this region are unknown (e.g., paired-end mapping, optical mapping)
- Variants for which we have basepair resolution for the breakpoints (e.g., sequencing)
When reporting variants there is a core set of data that will capture all the available information on your variant, including the degree of uncertainty present in the location of breakpoints. This data set includes:
start-stop coordinates: used to define events where breakpoints are known to basepair resolution. inner start-stop coordinates: used to define regions that are known to be affected by a variant, but do not define the actual breakpoints. The breakpoints lie outside of the defined region. outer start-stop coordinates: used to define the absolute outer boundary of a variation event but do not define the actual breakpoints. The breakpoints lie inside of the defined region. allele length: the length of the affected variant. For example, paired-end mapping may indetify a 5-kb deletion, but breakpoints are not known. Allele length does not have to be exact - approximations are acceptable, depending on the method.
The two structural variation archives (dbVar and DGVa) store data in a hierarchical fashion in an attempt to better capture the experimental process used to support variation features. There are basically two levels in the hierarchy, denoted by 'sv' and 'ssv'. Asserted variant regions (sv) are represent an author’s assertion about a variant region. Supporting variants (ssv) are meant to capture the experimental support for the asserted variant regions. ssv variant calls are children of sv calls. Additionally, sv calls can be merged with one another to add an additional layer to the hierarchy. There is no one standard for representing data of this complexity.
Your dbVar submission should report asserted structural variant regions (sv) and/or supporting variants (ssv).
In addition, dbVar works in close cooperation with the Database of Genomic Variants Archive (DGVa) at the EBI in Cambridge, UK. Unique identifiers ("accessions") are assigned to variants based upon which institution received the submission. Data are exchanged between the two resources on a regular basis. Identifiers, ‘sv’ and ‘ssv’, are prefixed with ‘n’ if the study was submitted to NCBI, or ‘e’ if it was submitted to EBI. Every dbVar variant will therefore have one of four (4) prefixes:
- nsv - asserted variant region submitted to NCBI
- nssv - supporting variant submitted to NCBI
- esv - asserted variant region submitted to EBI
- essv - supporting variant submitted to EBI
Likewise, study accessions are prefixed by 'nstd' if the study was submitted to dbVar, or 'estd' if it was submitted to DGVa.
Examples
Representing this information robustly is important for understanding the data within a particular study, for doing meta-analysis across studies, and for graphical rendering. Accurate reporting and use of structural variation (SV) data requires a familiarity with the methods used to generate the data and the lack of precision introduced by a given method. With the exception of some forms of sequencing data, precise variant breakpoints are rarely determined. The type and extent of uncertainty in defining breakpoints are an essential part of the variant data, and are required if the data is to be accurate and useful.
The following examples describe popular experimental methods currently in use to identify structural variants. Please read them carefully, as they explain exactly what data we expect to receive in your submission, depending on the experimental method(s) you used.
Current SV detection methods can be grouped into three types:
- Probe-based Methods (e.g., BAC array CGH, Oligo array aCGH, SNP arrays, etc)
- Mapping-based Methods (e.g., Paired-end mapping, Optical mapping, etc.)
- Sequencing-based Methods (e.g., Sanger sequencing, Next-gen sequencing, Sequence alignment, Read depth analysis, etc.)
If your method does not fall into one of the above categories, or if you used a combination of methods from more than one category, please use the information in the sections below as a guide. The goal is for you to provide complete variant descriptions, including the nature and extent of uncertainty in the location of variant boundaries.
Probe-based methods (BAC array CGH, oligo array aCGH, SNP arrays, read depth)
In this type of experiment, probes are arrayed across the genome (or genomic region) and signal intensity is used to identify regions that vary in copy number from what is expected. The density of probes at a given locus will determine resolution and therefore the extent of uncertainty. Because DNA between probes is not assayed by probe-based experiments, there is no way to capture the precise location of variant boundaries. Typically, the best one can do is identify the two probes that flank the breakpoin, one of which is affected by the variant, and the other of which is not. This necessarily results in “fuzzy” endpoints:
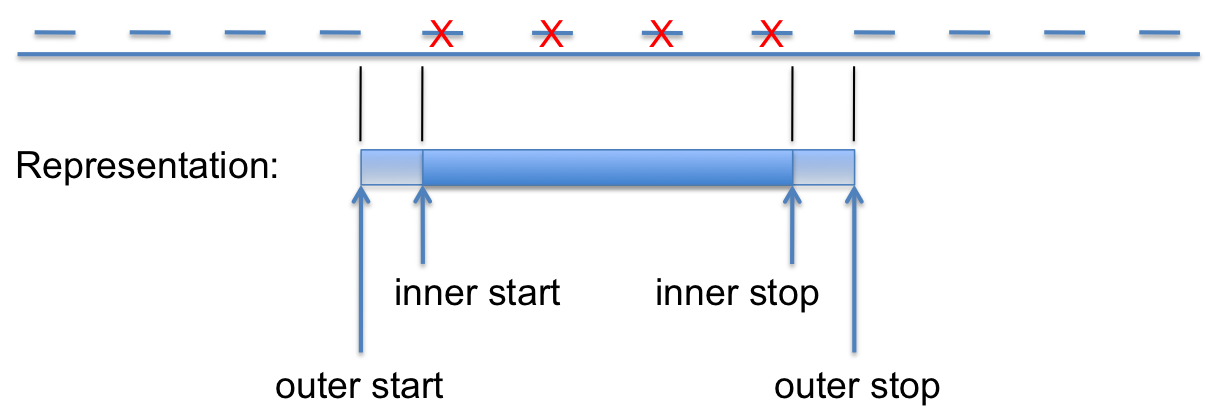
Figure 1: Uncertainty in Probe Data
In the example above, a group of probes are deleted (indicated by red Xs). The region between the inner_start and inner_stop coordinates is known to be deleted. Similarly, the region outside the outer_start and outer_stop coordinates is known to be present (i.e., not deleted, or unaffected by the variant). To capture all of the information it is necessary to report:
- inner_start = the 5’-most nucleotide of the first affected probe
- inner_stop = the 3’-most nucleotide of the last affected probe
- outer_start = the 3’-most nucleotide of the last unaffected probe preceding the variant
- outer_stop = the 5’-most nucleotide of the first unaffected probe following the variant
The same rules apply when there is an increase in probe intensity across a region (gain). However, there are important conceptual differences in the interpretation of gains and losses (see “An important note about Gains and Insertions” below.
If you are submitting SV data from a probe-based experiment (e.g., aCGH, SNP array) to dbVar, please include the following coordinates for each ssv, or supporting variant:
Included file 'expected_data-table1.inc' not found
Submissions based on probe-type experiments that lack either inner or outer start and stop coordinates are incomplete, but can be processed. The accurate identification and reporting of the extent of (un)certainty of regions that are known to be affected or unaffected are important aspects of the data. We ask that you submitt outer and inner coordinates wherever possible for probe-based experiments.
An important note about Gains and Insertions
- For probe-based gains, one does not know where in the genome the gained sequence is represented; it may be at the location represented in the array (e.g., a tandem duplication), or it may be elsewhere in the genome. However, like the deletion event, we do know which probes are part of the GAIN and can still provide precise information for the inner start/stop.
- One must distinguish between gains and insertions. Insertions can only be represented by mapping-based or sequencing-based data – they cannot be represented by probe data. A probe-based gain can be thought of as a duplicated (or “extra”) copy of known sequence content but unknown location. A mapping-based insertion can be thought of as extra sequence of unknown content but known, unknown or imprecise location. Gains are represented in probe data; insertions are represented in mapping data.
Mapping-based methods
In mapping based technologies (e.g., paired-end mapping, optical mapping), one knows with precision the outer boundaries between which the variant breakpoints must fall. In the case of paired-end mapping these are provided by paired-end sequences, while in optical mapping they correspond to restriction sites. The distance between outer_start and outer_stop coordinates can vary between 300bp and 40kb, depending on the method used. The size of the fragment also heavily influences the type and size of variant that can be detected.
In addition to outer start and outer stop, mapping data should provide the approximate size of the variant (allele_length). This is typically calculated by comparing the size of the experimental fragment to an expected size provided by a reference.
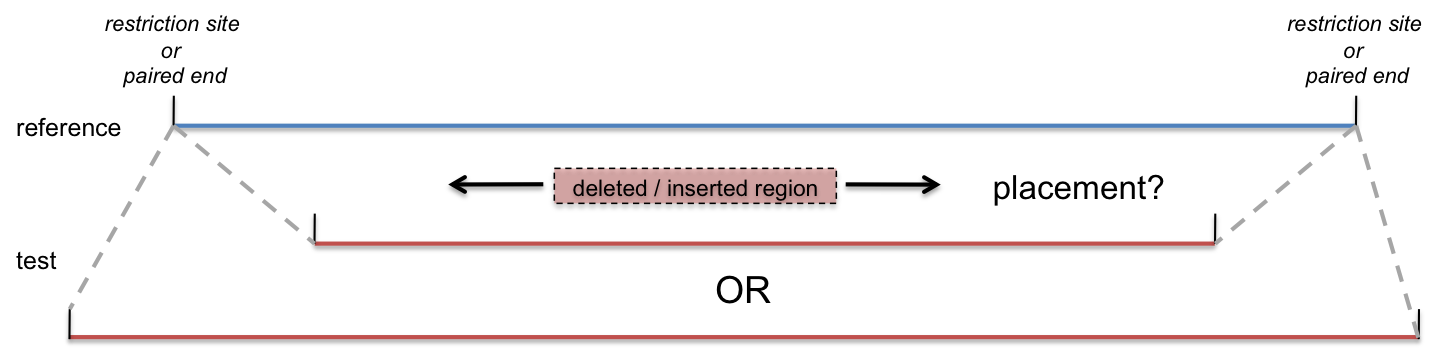
Figure 2: Uncertainty in Mapping Data
Note that the size of the variant (allele_length) often cannot be determined precisely. This lack of precision of the length of the variant is a separate issue from the fuzziness of the variant’s placement between the known endpoints (outer_start and outer_stop). It is not necessary to provide the degree of uncertainty in estimating allele length for every variant; rather, please include this resolution, and its rationale, in the method description.
The data we expect to receive with a mapping-based submission includes:
Included file 'expected_data-table2.inc' not found
Sequencing Based methods
In many cases, using either long reads or 2nd generation sequencing reads, breakpoint resolution of variants can be achieved. In such cases, we would expect the following data:
Included file 'expected_data-table3.inc' not found
However in many cases, 2nd generation sequencing may not give precise breakpoints and the user should give inner/outer starts as necessary.
Included file 'expected_data-table4.inc' not found
