Overview
What is Virus Variation Resource
Virus Variation (ViV) is an integrative, value-added resource designed to support the retrieval, display and analysis of large sequence datasets.
ViV includes expanded sequence data processing pipelines and analysis tools that together support selection and retrieval of nucleotide and protein sequences from several viral groups: influenza virus, Dengue virus, West Nile virus, Ebolavirus, MERS coronavirus, Rotavirus A and Zika virus.
New viruses, processing pipelines, tools, and other features are introduced regularly.
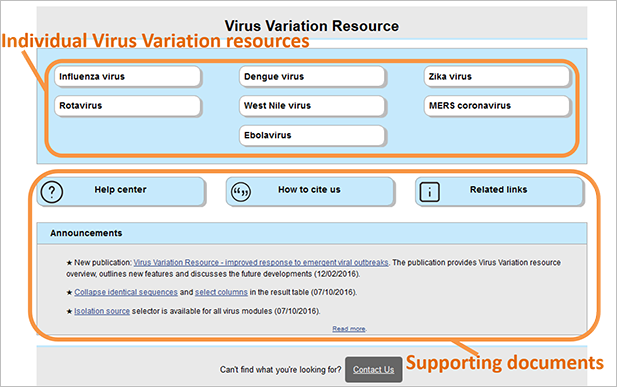
For Influenza virus resource we maintain a separate help documentation: Influenza Virus Resource help center .
How to cite The Virus Variation Resource
Virus Variation Resource - improved response to emergent viral outbreaks. Hatcher EL, Zhdanov SA, Bao Y, Blinkova O, Nawrocki EP, Ostapchuck Y, Schäffer AA, Brister JR. Nucleic Acids Res. 2017 Jan 4;45(D1):D482-D490. doi: 10.1093/nar/gkw1065. Epub 2016 Nov 28.
How to use Virus Variation Resource
Search interfaces for individual viruses (top panel) and help documents (bottom panel) can be accessed through the Virus Variation Resource home page .
Database search interface
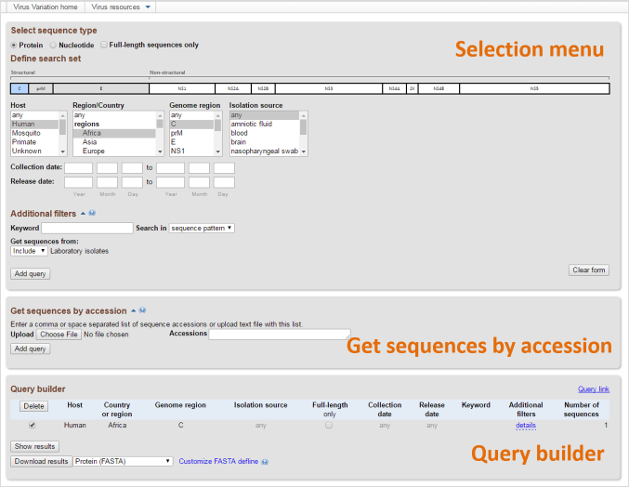
Find sequences of interest using a variety of criteria. It includes three main sections:
- Selection menu.
- Get sequences by accession (opened by pressing on "Get sequences by accession" heading).
- Query builder (opened if "Add query" button pressed).
Selection menu
This section is used to select nucleotide or protein sequences from the database using sequence and/or sample descriptors such genotype, genome region, disease, host, region/country, isolation source, collection date, and GenBank release date. Note that the selector options varies for different viruses. The menu supports multiple selections, so several proteins, hosts or geographic locations can be added to a single set of search criteria.
Additional filters
The Additional filters submenu is revealed by clicking the adjacent arrow icon. This section supports keyword searches for sequence patterns and strain names/definition lines. Filters for laboratory isolates, vaccine strains, and environmental samples are also included. The Rotavirus resource includes a secondary set of menus where users can select required segments/proteins and genotypes of selected segments/proteins. To learn more, please refer to Additional filters section on How To page.
Get sequences by accessions
This section supports retrieval of protein or nucleotide sequences using GenBank accession numbers separated by commas or spaces. You can also upload a file containing accessions. This function is especially useful if you want to add specific sequences to a dataset – like those used as a personal reference or found in a paper.
Query builder
After clicking the "Add Query" button, the search parameters and number of returned sequences are shown in "Query Builder.” Here results from individual searches can be combined by clicking the "Add Query" button after selecting new search criteria. Results from selected searches can also be downloaded in in a variety of formats using the “Download results” button. Please refer to How to use Database Query Interface section to learn more.
Supporting tools and displays
Search results interface
Search results can be viewed in the results interface on a separate page by clicking the "Show results" button under the Query Builder table.
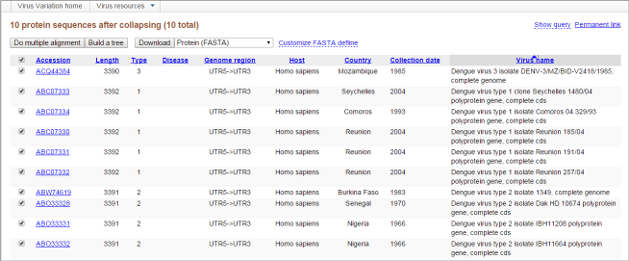
The search result page displays retrieved sequences from search sets along with several sortable metadata columns and supports selection of individual sequences for download or further analysis. Sequences can be downloaded, and it is possible to analyze them using the multiple sequence alignment or tree building tool integrated to the database. Please learn more in How to use Results View section.
Multiple sequence alignment viewer
Multiple sequence alignments can be calculated from selected protein or nucleotide sequences using MUSCLE .
Clicking the "Do multiple alignment" button creates an alignment from selected sequences and displays the alignment in a new page in a viewer based on the NCBI Genome Workbench multiple sequence alignment viewer. This viewer includes a variation histogram above the alignment as well as a feature table that highlights mature protein boundaries and other important sequence features. Please, learn how to use multiple alignment viewer here .
Tree Viewer (TV)
Clustering or phylogenetic trees can be calculated from selected protein or nucleotide sequences.
Clicking the "Build a tree" button located on both the results and alignment pages generates a tree from selected sequences. The tree is displayed in a second page using the NCBI Tree Viewer. This viewer provides a variety of options for highlighting isolates within a tree, and trees can be downloaded in PDF, ASN1 (text and binary), Newick, and Nexus formats.
To learn what you can do with a tree using TV, please refer to How to use Tree Viewer section.
