About Identical Protein Groups (IPG)
Background and Scope
Searching within the Protein resource can be daunting because an overwhelming number of records may be returned by a text search. The Identical Protein Groups (IPG) resource makes it easier to find protein information by searching against groups of protein records where each group represents a unique protein sequence. This reduction in the database size makes it easier to find information by text search. IPG report pages, which were already available as an alternate display from the Protein resource, identify members of the protein group and include additional information such as corresponding nucleotide coding sequence regions, protein names, and organism names. Now these Identical Protein Reports can be found in a single searchable resource, Identical Protein Groups (IPG), available at https://www.ncbi.nlm.nih.gov/ipg/.
Advantages to searching in the IPG resource rather than in the Protein resource include:
- Smaller search results sets are returned, since there will be only one result for each unique protein sequence. Having smaller sets will enable the investigator to more quickly identify a protein sequence of interest.
- Identical protein groups include protein accessions from GenBank, RefSeq, SwissProt, PDB, and other sources. Protein groups can be searched using protein accession numbers and text searches, such as for a protein name or by taxonomic terms.
- The IPG report highlights in its summary a ‘best’ RefSeq or SwissProt example accession, when available, and its protein name so investigators may choose to focus on those sets.
- Searches can be made against all GenBank protein translations including those from annotated WGS genomes which will not be included in the Protein resource in the future. All GenBank proteins are in the Identical Protein Reports and can be found via the IPG resource.
- The lowest common taxonomic group name (e.g., mammals, or g-proteobacteria) is indicated for each unique protein and can be used to identify highly conserved proteins. However, some apparent divergence may have alternate causes such as sample mix-up, misclassification or contamination in one of the samples, or may reflect that the protein is found in a mobile element (eg, from a phage or virus) or is a selectable marker (eg, chloramphenicol acetyltransferase).
- The number of identical proteins in each group is presented, so one can identify more supported proteins where the identical protein has been observed many times.
- IPG reports include a downloadable table that maps protein accessions, nucleotide record (CDS) coordinates, and taxonomic information. This is especially important for RefSeq bacterial proteins which use a distinct data model whereby one RefSeq protein (the non-redundant WP_ accessions) can be annotated on many distinct genomes (see https://www.ncbi.nlm.nih.gov/refseq/about/nonredundantproteins/#identicalprotein).
Record Description
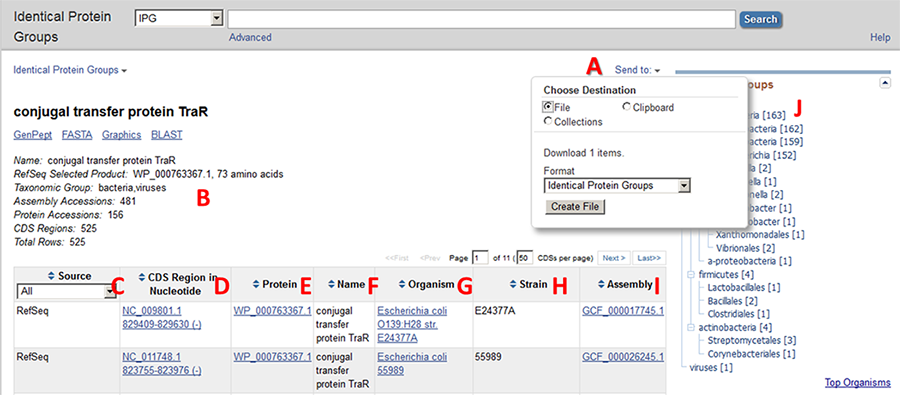
The top section of a group report is shown below. The report includes the following information and features:
A: Report contents can be downloaded as a tabular file for local use.
B: The top section of the report page shows the product name of the group being viewed. Beneath that is a summary about this group:
- the product name that is found on the preferred protein accession
- the automatically selected preferred protein accession ('RefSeq Selected Product') and the protein length
- the lowest common taxonomic node(s) of the proteins in the group
-
the number of:
- assembly accessions
- protein accessions
- CDS regions
- total rows
- For GenBank and RefSeq proteins, each protein exists as annotation from a nucleotide record so each protein accession has a corresponding coding region. However, the Identical Protein Groups resource includes proteins that are not just annotations; these protein-only accessions come from SwissProt, PDB, and patent (PAT) records which do not have corresponding nucleotide coding regions.
C: The database source is indicated. This may be RefSeq, INSDC (ie, GenBank and the other members of the International Nucleotide Sequence Database Collaboration), SwissProt, PDB, PIR, or Patent.
D, E: The nucleotide accession and coordinates (D) correspond to the annotated CDS feature which contains a cross-reference to the protein accession (E). The link on the nucleotide coordinates navigates to the Nucleotide database to display the annotated CDS region in GenBank format. The link on the protein accession number returns that protein record in GenPept format.
F: The product name of that protein accession.
G, H: The Organism and Strain columns report information corresponding to the NCBI Taxonomy identifier that is annotated on the Nucleotide record. If a nucleotide record is not available then the values reported correspond to data found on the protein record.
I: The Assembly column reports the assembly accession in which that coding region appears, when the corresponding nucleotide record is part of a genome assembly
J: The Taxonomic Groups section presents a taxonomic tree of the organisms from column G for this group. Clicking on a particular branch of the tree will limit the report to only those proteins in that taxonomic lineage. Clicking on the “Show all” link that appears at the top of the table returns the full list.