Using the Sequence Viewer Embedding API
- Introduction
- Embedding Sequence Viewer in your code
- Configuring the View
- Automatic and programmatic initialization
Introduction
The NCBI Sequence Viewer is an interactive visualization tool for displaying molecular biology information (sequence data plus annotation). It offers the user a fully interactive context which includes:
- Custom set of tracks (e.g. gene annotations)
- Zooming to the area of interest
- Setting markers on genes or sequence positions to illustrate the focus of your research
- Tooltips that show more detailed information and include hyperlinks to related resources.
An embedded viewer is better than a link to a viewer, because a link will take your user from your page to an external resource. The embedding page can use many layers of API from simple inclusion of the viewer up to a fully integrated web application.
For step by step description of embedding the NCBI Sequence Viewer into your page please watch the instructional video:
Additional documentation
You can also familiarize yourself with other existing documentation:
- Examples files (Example1-4)
- Embedding API Manual
- Embedding demos (shows examples of embedded SViewer)
- URL parameters
- Sequence Viewer event listening demo
- General NCBI Sequence Viewer documentation
Examples of NCBI Sequence Viewer embedded on NCBI pages
You can show Sequence Viewer on your own site configured by parameters including the anchor sequence, position and zoom level, set of tracks, markers, and more.
Embedding Sequence Viewer in your code
This code example can be examined in the file Example1 from the archive. It demonstrates automatic initialization of the DIV with Sequence Viewer.
Code for embedding the Sequence Viewer. One line in the HEAD section:
<script type="text/javascript" src="https://www.ncbi.nlm.nih.gov/projects/sviewer/js/sviewer.js"> </script>
Code to instantiate the specific view in the BODY of your HTML file:
<div id="mySeqViewer1" class="SeqViewerApp" data-autoload> <a href="?embedded=true&appname=testapp1&id=NC_000001"></a> </div>
Details to pay attention to:
- Class of the DIV with Sequence Viewer should be set to a fixed value, “SeqViewerApp”.
- “Data-autoload” ensures that this DIV will be found and automatically instantiated by Sequence Viewer code.
- Always fill “appname” field with the name of your application, preferably prefixed with the name of your organization. There are no formal rules for the name, but it should not contain spaces (you can use underscores), and should not have special letters for TM, Copyright etc. This name allows us to provide you with better service and allocate necessary resources.
- Use entity for ampersand separating the parameters instead of just ‘&’. While usage of ‘&’ is tolerated by modern browsers it is prone to accidental errors.
Configuring the View
The easiest way to configure the view is to borrow the config line from an existing instance of the Sequence Viewer. Let's configure a view (set of tracks, zoom, position, markers) and embed it on your page.
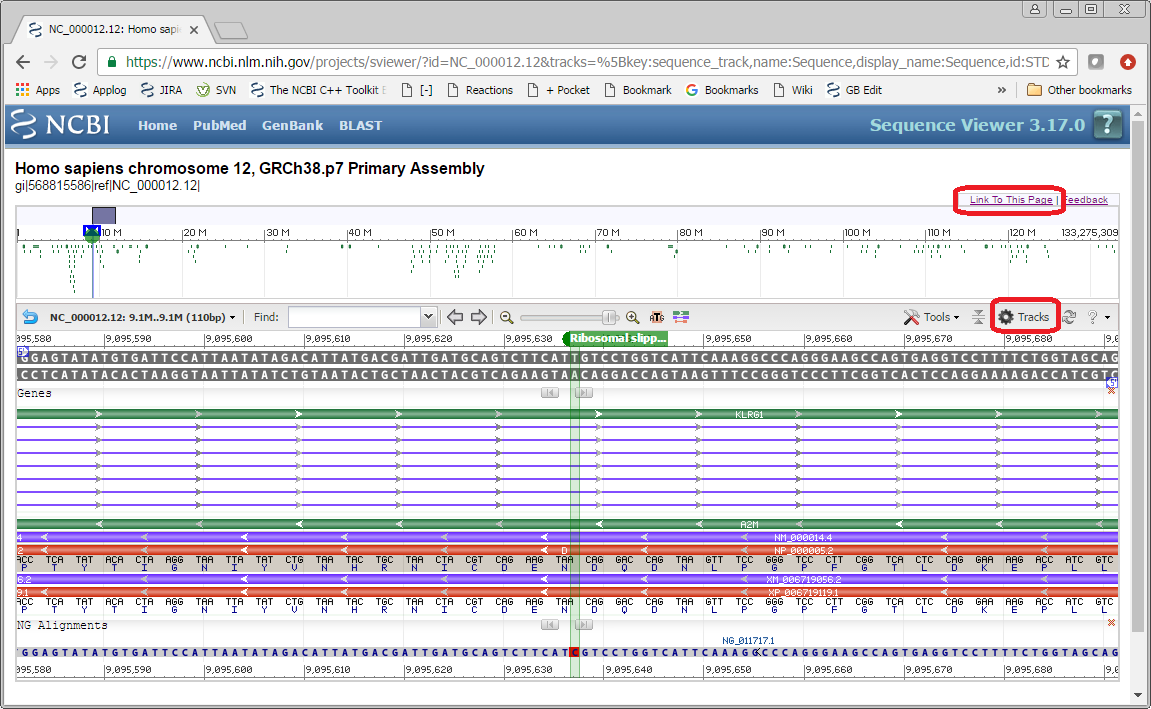
Open the embedding example of genomic to refseq mismatch
You can now configure the view the way you like. You can change the zoom, coordinates shown, or add or remove tracks using the Tracks menu.
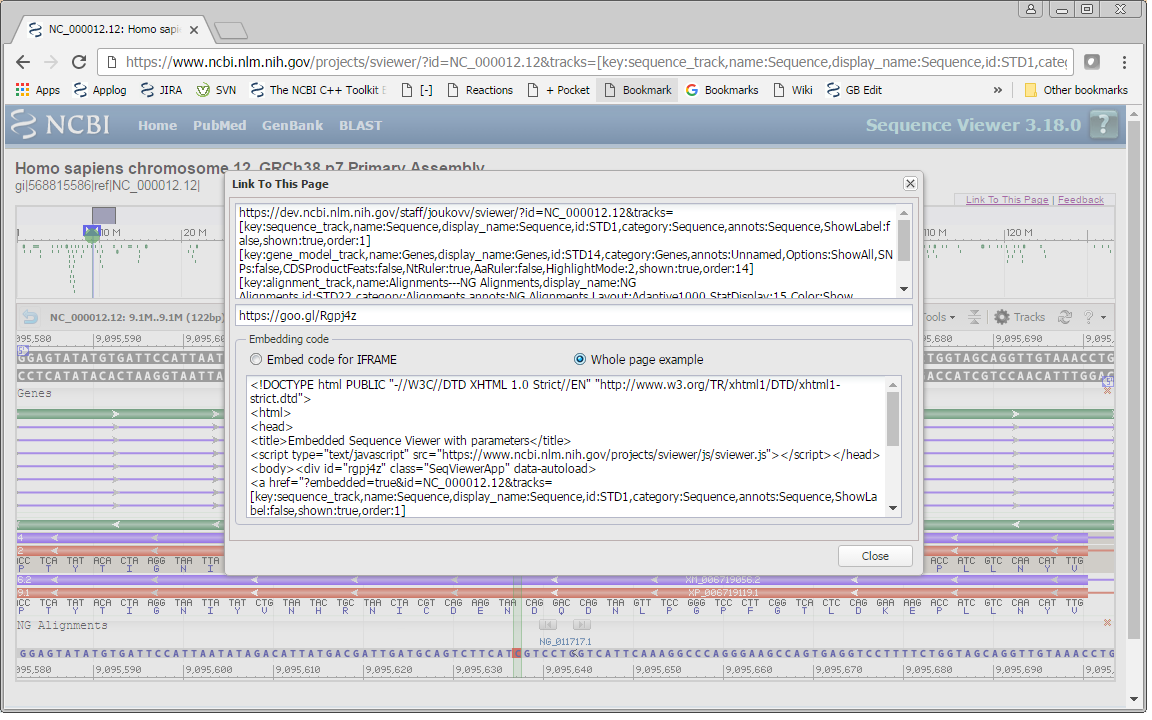
After you’re satisfied with your view, click on the Link To This Page button in the right top part of the page. The window with three text areas appears. You can copy text from these areas and use it in your code.
The first two areas contain a link that you can use to save or share the exact view you configured. The second one is just a short link to the view.
The third box contains code for embedding this view on your page. You have the choice of two types of code. The first option, ‘Embed code for IFRAME’, can be used as is to embed the Sequence Viewer by itself as an IFRAME. The second option,‘Whole page example,’ can be used to embed the Sequence Viewer as a part of your page. This option has both the script in the HEAD element and DIV element with parameters. Check out Example2, which contains the code with “appname” parameter replaced with our own application name.
Automatic and programmatic initialization
Example2 used automatic initialization when you just supply the parameters in an HTML element and Sequence Viewer does the job.
Sometimes the exact parameters are unknown beforehand and you need to supply them as a result of some operation, e.g. choice of your users. You can show a list of human chromosomes (for the sake of the demo, short list of first 5 chromosomes), put them in a 'SELECT' element, and activate them upon user choice. See Example3 for full text of this code.
Adding publicly available data
Your data can be displayed alongside NCBI sequence or tracks in the Sequence Viewer. This data should be located on publicly available FTP or HTTP service and in a formats understood by Sequence Viewer. The list of formats includes BED, FASTA, GFF2, GFF3, GVF, GCF, Wig, VCF, etc.
Add url=path_to_your_data parameter to your embedding code to use your data. Say you have a BED file and you want to display it on the Human chromosome 1, NC_000001.10.
Let’s view at the region around 115M bases – add parameter for view, v=105M:125M.
The full URL to view is https://www.ncbi.nlm.nih.gov/projects/sviewer/?id=NC_000001.10&url=ftp://ftp.ncbi.nlm.nih.gov/toolbox/gbench/samples/UCSCGenes_exon20.bed&v=105M:125M
You can observe the embedded view in Example 4.
If you have any questions please contact us.
Table of Contents
- Sequence Viewer application
- Documentation Home
- General
- Help
- Interface
- Tutorials
- Manuals
- Demo pages
- Related Resources