Hsp90 is an essential and ubiquitous molecular chaperone that is required for the proper folding of a set of client proteins at a late stage in their folding process. In eukaryotes, cytoplasmic Hsp90 is absolutely essential for cell viability under all growth conditions. The functional cycle of the Hsp90 system requires a cohort of cochaperones and cofactors that regulate the activity of this chaperone. Hence, Hsp90 function is highly complex; in order to understand that complexity, several groups have attempted to map out the interaction network of this chaperone in yeast and mammalian systems using the latest available proteomic and genomic tools. Interaction networks emerging from these large scale efforts clearly demonstrate that Hsp90 plays a central role effecting multiple pathways and cellular processes. In yeast Saccharomyces cerevisiae, Hsp90 was shown to interact directly or indirectly with at least 10% of the yeast ORFs. The systematic application of large scale approaches to map out the Hsp90 chaperone network should allow the determination of the mechanisms employed by this chaperone system to maintain protein homeostasis in the cell.
Introduction
Hsp90 is an abundant and essential molecular chaperone present in all eukaryotic cells. Hsp90 has been implicated in mediating the folding of a specific set of substrate proteins, which are typically called ‘clients’. Hsp90 clients are thought to interact with the chaperone in a nearly native state at the late stage of de novo folding;1 alternatively, Hsp90 can also enhance the rate of reactivation of heat denatured proteins under stress conditions.2 Although no common sequence or structural features have yet been identified for Hsp90 clients, however, many of the early clients that were identified fell into two functional classes: transcription factors3 and protein kinases.4 These early observations suggested the involvement of Hsp90 in modulating signaling pathways. Hsp90 is, therefore, considered to be particularly important for cell cycle control, organism development, and response to environmental stresses.5,6 Hsp90 has become a novel anti-cancer drug target, and there are several drugs that specifically bind to Hsp90 and are in clinical trials.7,8
Hsp90 is a dimeric protein consisting of three domains: an N-terminal ATP-binding domain, a middle region, and a C-terminal domain involved in homodimerization. The N-terminal domain has a unique fold termed the Bergerat fold.9 Although the ATPase activity of Hsp90 is weak, ATP binding and hydrolysis is crucial to the chaperone function.10 Several drugs that have been designed to target Hsp90, such as geldanamycin, bind in the ATP pocket at the N-terminal domain. In eukaryotes, the C-terminal domain of cytosolic Hsp90 ends with a conserved EEVD motif. This motif has been shown to be essential for the interaction of cytosolic Hsp90 with cofactors that contain α-helical tetratricopeptide repeat domains (TPR).
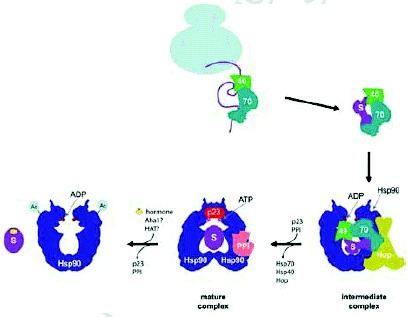
The Hsp90 functional cycle has been intensively investigated, however, due to the complexity of this chaperone system and the diversity of the clients, a full understanding of the molecular aspects of the cycle is still lacking. It is known that the activity of Hsp90 requires the involvement of other chaperone systems such as Hsp70 and Hsp40 as well as several cofactors (Fig. 1). The Hsp90 functional cycle has been well-studied for the case of steroid hormone receptors (Fig. 1).3,11 It is thought that the monomeric inactive receptor is initially recognized by the Hsp70/40 system and, subsequently, transferred to Hsp90. The formation of a complex between Hsp70 and Hsp90 is mediated by the scaffold protein Hop/p60 (Sti1 in yeast). In yeast, Sti1 has been shown to be an activator of the Hsp70 ATPase and an inhibitor of Hsp90 ATPase, although mammalian Hop does not seem to have such an effect.12,13 Subsequently, other cyclophilins (e.g., yeast Cpr6 and Cpr7) and immunophilins (e.g., yeast Cns1) as well as the cochaperone p23 (Sba1 in yeast) interact with the Hsp70/40-Hop-Hsp90-substrate complex resulting in the release of Hsp70/40 and Hop. p23 stabilizes the Hsp90-receptor complex in the ATP state. Upon arrival of the hormone into the cell, the receptor binds the hormone and is released from Hsp90 upon hydrolysis of the chaperone-bound ATP to ADP; the receptor is then active as a dimer and translocates into the nucleus. The ATPase activity of Hsp90 is enhanced by the cofactor Aha1/Hch1.14,15 It has also been demonstrated that Hsp90 activity can be regulated by phosphorylation by the phosphatase Ppt116 in yeast and by acetylation/deacetylation by an unknown HAT and by HDAC6 in mammalian cells, respectively.17,18 Several other Hsp90 cofactors are also known, such as Cdc37 and CHIP, and they affect the chaperone activity in different manners. It is generally thought that different Hsp90 cofactors target the chaperone to different sets of substrates.
Recently, several systematic genomic and proteomic approaches were employed in order to map out the Hsp90 molecular interaction network. These approaches provide a global view of the influence of the Hsp90 system on multiple processes and complexes in the cell. They also shed more light on the mechanism of Hsp90 function. The description of these approaches follows below.
Mapping the Hsp90 Physical Interaction Network
The Hsp90 physical interactors represent a class of proteins that either directly interact with Hsp90 or that are part of complexes that interact with Hsp90. Most of these Hsp90 physical interactors are thought to mainly include Hsp90 cofactors and cochaperones as well as Hsp90 clients. Two different methods have been employed to map out such interactors: (1) pull-down or immunoprecipitation of the chaperone to isolate Hsp90-containing complexes followed by mass spectrometry to identify proteins in those complexes, and (2) two-hybrid method to identify proteins that, predominantly, directly bind to Hsp90.
Isolation of Hsp90 Complexes Followed by Mass Spectrometry
Isolation of Hsp90 complexes followed by mass spectrometry to identify components of the complexes is an effective and straight forward approach to explore the Hsp90 interactors. Matrix assisted laser desorption ionization time of flight (MALDI-ToF), tandem electrospray mass spectrometry (MS/MS), or liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) have been extensively used to identify single proteins or protein mixtures19 in gels or in solution. While protein identification might be robust, this approach greatly depends on the quality of purified complexes.
This method has been used to isolate Hsp90 complexes either by affinity pull-down of tagged Hsp90 in yeast or by coimmunoprecipitation of Hsp90 from mammalian cells. Zhao et al20 introduced a TAP-tag to endogenous Hsp90 in yeast, while keeping the gene under the control of its own promoter. The TAP-tag is a tandem affinity purification tag containing two IgG binding domains from Protein A and a calmodulin binding peptide motif separated by a tobacco etch virus (TEV) protease cut site.21 Yeast has two virtually identical HSP90 genes, one termed HSP82, which is heat shock induced, while the other is termed HSC82, which is constitutively expressed. The proteins are localized in the cytoplasm. Strains were constructed in which endogenous HSC82 or HSP82 were C-terminally TAP-tagged, in addition, a third strain was constructed in which HSP82 was deleted, while HSC82 was N-terminally TAP-tagged. Hsp90-containing complexes were then isolated on IgG beads, released using TEV protease, further purified on calmodulin beads in the presence of calcium, and then released by addition of EGTA to chelate the calcium. This approach allowed the isolation of native protein complexes that were purified to more than 106 fold to virtual homogeneity. Since the EEVD motif at the C-terminus of Hsp90 is essential for the interaction of Hsp90 cofactors with the chaperone, most of the useful complexes were obtained from the third strain in which HSC82 was N-terminally TAP-tagged and HSP82 was deleted. It should be noted that, in this approach, the addition of the tag might affect Hsp90 activity. Indeed, the yeast strain with N-terminally TAP-tagged Hsp90 had slight temperature sensitivity. Isolated proteins were identified by mass spectrometry and some ‘hits’ were then verified using reverse pull-downs in which the identified proteins were TAP-tagged and pulled-down.
To identify Hsp90 interactors in mammalian systems, Falsone et al22 used a commercially available anti-Hsp90 antibody to immunoprecipitate Hsp90 complexes from human embryonic kidney (HEK293) cells. One of the hits was verified by determining that its levels were sensitive to the inhibition of Hsp90 by the drug geldanamycin. It should be noted that in the human genome there are seventeen genes which have been detected to belong to the Hsp90 family.23 Six of these genes were recognized as functional, while the other eleven genes were classified as pseudogenes. These Hsp90s are present in different compartments of the eukaryotic cell further complicating the studies on mammalian Hsp90.
Two Hybrid Screens
The yeast two-hybrid (2H) system is one of the most widely used methods to screen or confirm protein-protein interactions.24 The basic principle of this system is that a protein X, the bait, is fused to a DNA binding domain (DBD), which is commonly derived from the Gal4 or LexA transcription factors. Protein Y, the prey, is fused to a transcription activation domain (AD). If proteins X and Y interact, then AD and DBD are brought close to each other, triggering the activation of a downstream reporter gene.
Despite the popularity of this method, there are several drawbacks that have to be considered. The interactions have to occur in the nucleus and a nuclear targeting sequence is present in the constructs used. In this regard, yeast Hsp90 is predominantly cytoplasmic and it has been reported that the chaperone has a cytoplasmic localization signal in its sequence.25 The nuclear localization signal in the DBD domain may not be strong enough to bring the large Hsp90 to the nucleus and this is indeed observed for full-length yeast Hsp90 (R. Zhao and W.A. Houry, unpublished results). Also, the DBD-Hsp90 fusion might not be fully active. It has been reported that a C-terminal Hsp90-Gal4 fusion can support yeast growth better than an N-terminal Gal4-Hsp90 fusion.26 Also, the interaction between Hsp90 and its client proteins is probably transient and the interaction might not be strong enough to activate the downstream reporter gene. Nevertheless, this approach has been successfully used by the Piper and Houry groups20,27,28 to map out the Hsp90 2H interaction network using an ordered array of about 6000 yeast ORFs fused to Gal4 activation domain that was created by Uetz et al.29
Mapping the Hsp90 Genetic Interaction Network
Hsp90 genetic interactors represent a class of proteins that are involved in a pathway where Hsp90 also plays key functions in parallel. Hsp90 genetic interactors may or may not directly interact with the chaperone, but both, the chaperone and its genetic interactor, are required for the successful function of a particular essential cellular process or pathway, and consequently, for cell viability. The large scale identification of Hsp90 genetic interactors has been successfully used for yeast Hsp90.20
Synthetic Genetic Array
Synthetic genetic array (SGA) analysis is based on the principle of synthetic lethality developed for the budding yeast Saccharomyces cerevisiae.30 For this organism, about 80% of the open reading frames are nonessential.31 Tong et al30 developed automated methods to screen for synthetic lethality of a given knockout strain against the systematic gene deletion array32,33 consisting of about 4700 haploid strains each of which has a marked deletion of one nonessential gene. A knockout strain is mated with the deletion array to eventually obtain strains with double deletions. If these strains are sick or not viable, then the two deleted genes are said to be genetic interactors. Putative hits can be verified by tetrad analysis or random spore analysis. The detailed method and its application has recently been reviewed in reference 34.
This screen is ideally designed for a query of nonessential gene; however, it is also suitable for an essential gene query if a conditional knockout, such a temperature sensitive mutant of the essential gene, is available. Hsp90 is essential in all eukaryotic cells. A number of temperature sensitive yeast Hsp82 mutants such as Hsp82(G170D), Hsp82(G313S), and Hsp82(E381K) have already been identified and characterized.35 An SGA analysis of Hsp90 interactors has recently been carried out using a strain in which hsc82 is deleted and HSP82 is rendered temperature sensitive by introducing the G170D mutation.20 The screen was carried out at the semi-permissive temperature of 35°C.
Chemical Genetic Screen/Drug Screen
The chemical genetic screen or drug screen is based on the principle of synthetic lethality and takes advantage of the unique bar codes introduced to mark each deletion mutant in the systematic deletion array.32 These bar codes can be amplified using common primers. In this method, the bar-coded haploid deletion mutants of the nonessential genes31 are pooled and grown in liquid culture containing an Hsp90 specific drug inhibitor, geldanamycin, to challenge Hsp90 function. Strains containing deleted genes that are synthetic lethal or synthetic sick with Hsp90 will die or grow slower compared to other strains. The same experiment is repeated in the absence of the drug. The common primers are then used to amplify genomic DNA from the pooled strains and the resultant products are hybridized onto specific DNA oligo array chips. Strains that do not grow or that grow slowly in the presence of the drug will not give a signal or will give a lower signal when compared to the signal obtained in the absence of the drug. The hits obtained from applying this technique should closely overlap with the hits obtained from the SGA analysis as both methods depend on the same principle of synthetic lethality. Zhao et al20 employed this method to determine yeast Hsp90 genetic interactors using the drug geldanamycin (geldanamycin screen or GS).
The Hsp90 Interactome
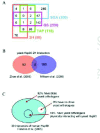
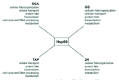
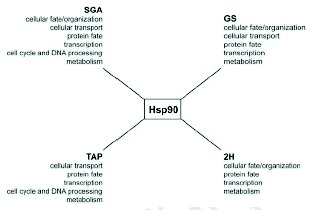
Yeast is currently the ideal system for large-scale genetic and proteomic studies due to the ready availability of many tools for such studies in this organism. Figure 2A gives an overview of the number of hits obtained using the four different methods described above for yeast Hsp90 as reported by Zhao et al.20 The Hsp90 interactors (over 600 ORFs) represent about 10% of the yeast proteome and are distributed among multiple cellular functional categories (Fig. 3) suggesting that Hsp90 is involved in many processes. One feature that stands out in Figure 2A is that there is little overlap among the different datasets obtained using the different methods. This reflects the experimental differences between the methods used as each method favors certain types of interactions. The best overlap was obtained between the SGA and the geldanamycin screen (GS). A total of 451 Hsp90 genetic interactors were identified from the two screens with an overlap of 49 hits. These 49 hits represent a high fidelity dataset that could then be pursued further for more detailed biochemical studies. The overlap is considered to be reasonable.
On the other hand, the physical interaction screens gave a total of 208 hits with an overlap of only 10 hits between the TAP and 2H screens (Fig. 2A), which is less than expected. The differences might arise because the 2H screen predominantly detects binary interactions, while the TAP method detects interactions between the bait and a complex of polypeptides that need not directly bind to the protein bait. Hence, the 2H screen might reveal weaker interactions than the TAP screen.
It is interesting to note that no protein was simultaneously detected by the four methods used and that only Cpr6, Cpr7, Sti1, and Mon1 were detected by three of the four screens. Cpr6, Cpr7, and Sti1 are all well-established Hsp90 cofactors, while the interaction of Hsp90 with Mon1 has not been reported before; therefore, Mon1 is likely an Hsp90 substrate. Mon1 is poorly studied; however, it is known to form a complex with another protein termed Ccz1 that binds the vacuolar membrane and is involved in homotypic vacuole fusion.36,37 This might implicate Hsp90 in vacuolar trafficking.
There were 22 proteins that were found to have the unique property of interacting with Hsp90 physically and genetically (Fig. 2A).20 These proteins are either important cofactors of the Hsp90 chaperone system that become essential when the chaperone's activity is reduced. Alternatively, these proteins are substrates of Hsp90 that physically interact with the chaperone. In this scenario, Hsp90 might affect directly or indirectly the folding of two proteins X and Y and is found to physically interact with X (one of the 22 proteins). X and Y have to be genetic interactors such that strains deleted of both the X and Y genes are not viable. Hence, when Hsp90 function is compromised, X and Y will not fold properly and synthetic lethality results when X is deleted in a strain background with reduced Hsp90 activity. Identifying two proteins to be physically as well as genetically interacting is not expected to be common and might be a peculiar property of chaperones.
2H screens for yeast Hsp90 have also been carried out by Millson et al.27,28 Initially, the group used full length Hsc82 as bait; however, even with DBD domain fused to the C-terminus of Hsc82, which is reported to have similar activity as the wild type chaperone, only four interactors were identified including the known Hsp90 cofactors Aha1 and Hch1.27 To detect Hsp90 clients that probably only weakly interact with Hsp90, Millson et al repeated the 2H screen using a mutant Hsp82(E33A). This form of Hsp90 is barely functional and is thought not to proceed to the final maturation step in the chaperone functional cycle due to failed ATP hydrolysis, which in turn is thought to hold the client proteins tighter rendering them easier to detect by 2H. A total of 177 genes including some known Hsp90 cochaperones and substrates were identified using this approach.28 In this regard, it is surprising to find that one-third (59 out of 177) of the ORFs identified by Millson et al28 are putative membrane proteins, putting into question the validity of this screen.
In comparison, Zhao et al20 used full length yeast Hsp82 as well as the different domains of Hsp82 as baits. This approach might be better in identifying physiological substrates and cofactors of Hsp90 as compared to the use of an inactive mutant of the chaperone.28 A total of 90 Hsp90 interactors were identified20 mostly by binding to Hsp90 domains as only 8 interactors were found to bind to full length Hsp82. In this screen, only 6 proteins out of the 90 are putative membrane proteins.
Eight proteins overlap between the 2H datasets of Zhao et al20 and Millson et al28 (Fig. 2B). These proteins are: Aha1, Arr3, Bsd2, Cns1, She3, Slt2, Sor2, and Tah1. This low overlap might reflect differences in experimental conditions and baits used. The 8 proteins are probably robust interactors of Hsp90. In fact, it is already known that Aha114,15 and Cns138 are Hsp90 cofactors that interact with Hsp90 in vivo and in vitro. Furthermore, Zhao et al20 and Millson et al28 verified the interaction of Hsp90 with Tah1 and Slt2, respectively, using in vitro assays.
The high throughput analysis of mammalian Hsp90 interactors is limited. Physical interactors of mammalian Hsp90 in HEK293 cells were identified using a simple procedure of immunoprecipitation of Hsp90 complexes and the identification of proteins in those complexes using nano-LC-MS/MS.22 Falsone et al22 identified 39 proteins as Hsp90 interactors (Fig. 2C). These proteins include metabolic enzymes, ribosomal subunits, and components of the cytoskeleton. However, many of the known Hsp90 interactors were not detected. The identified proteins are mostly housekeeping proteins and are abundant in the cell. This reflects the nature of the coimmunoprecipitation method; that is, most of the weak and transient interactions, especially with low abundance proteins, are missed. However, even for the short list of human Hsp90 physical interactors obtained, it is noticed that 36 out of 39 of the interactors (except for P04075, Q9NR45 and Q96KM8) have close yeast orthologues (Fig. 2C). Some of these orthologues (yeast Act1, Mdh3, Tdh3, and Yer156c which correspond to human P63261, P00338, P00354, and Q9HB07, respectively) were also found to physically interact with the yeast Hsp90 by TAP20 or 2H28 screens (Fig. 2C). This may suggest that not only the Hsp90 chaperone machinery itself, but also the core Hsp90 chaperone network may be somewhat conserved across organisms.
Perspectives and Future Directions
The integrated approach used by Zhao et al20 could also be used for other chaperone systems in yeast. The ultimate goal of mapping the chaperone interaction networks in the cell is to determine the in vivo mechanisms that govern protein homeostasis. In other words, if these screens are repeated often enough and if high fidelity data sets are obtained using multiple methods under different conditions, can a set of rules be eventually derived that would allow us to a priori predict how a given protein will fold inside the cell: what chaperone systems are recruited to help in the folding process and what types of interactions are important for proper de novo folding?
None of the current reported large-scale interaction studies on Hsp90 have been able to find a consensus sequence or structural motif that is preferentially recognized by this chaperone. This is in contrast to other simpler systems such as Hsp60 in E. coli (GroEL) in which preferential binding of GroEL to αβ folds or TIM barrels has been suggested.39,40 This seems to point to the functional complexity of the Hsp90 system and the fact that substrate specificity is probably dictated, to a certain extent, by the cofactors and cochaperones of Hsp90. Therefore, mapping the chaperone interaction networks must be carried out using multiple genetic and proteomic screens and varying experimental conditions in order to obtain a comprehensive view of the cellular protein folding landscape and in order to minimize false negative and false positive hits. For example, even with the integrated proteomic and genomic approach employed by Zhao et al,20 some well-established Hsp90 interactors were missed such as Sba1/p23 and Hap1. The interaction between Hsp90 and Sba1/p23 is stable only in the presence of ATP. In another example, physical interactions with membrane proteins are usually difficult to detect. The application of a recently developed split-ubiquitin membrane yeast two-hybrid system might allow the detection of such interactors.41,42
With the development of new techniques, the integrated approach used by Zhao et al20 could also be applied for mammalian Hsp90: genetic interactors can be revealed using RNA interference technology, while physical interactors can be studied using 2H and pull-down/mass spectrometry methods. Furthermore, investigating Hsp90 interactors in a particular tissue or organ, such as human liver or kidney, might be more meaningful than a whole genome-wide screen of Hsp90 interactors. The interactions identified in these organs or tissues could reveal site-specific roles of Hsp90. Finally, quantitative proteomics has been used as a sensitive method to monitor the dynamics of a proteome.43 Future studies should concentrate on examining the change of the Hsp90 interactome under different stress conditions. Hence, the hope is that future chaperone networks will evolve from static pictures to dynamic entities.
Acknowledgements
Work in the author's laboratory is supported by research grants from the Canadian Institutes of Health research, Natural Sciences and Engineering Research Council of Canada, and the National Cancer Institute of Canada.
References
- 1.
- Buchner J. Hsp90 & Co. A holding for folding. Trends Biochem Sci. 1999;24(4):136–141. [PubMed: 10322418]
- 2.
- Nathan DF, Vos MH, Lindquist S. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci USA. 1997;94(24):12949–12956. [PMC free article: PMC24244] [PubMed: 9371781]
- 3.
- Pratt WB, Galigniana MD, Morishima Y. et al. Role of molecular chaperones in steroid receptor action. Essays Biochem. 2004;40:41–58. [PubMed: 15242338]
- 4.
- Sreedhar AS, Soti C, Csermely P. Inhibition of Hsp90: A new strategy for inhibiting protein kinases. Biochim Biophys Acta. 2004;1697(1-2):233–242. [PubMed: 15023364]
- 5.
- Young JC, Moarefi I, Hartl FU. Hsp90: A specialized but essential protein-folding tool. J Cell Biol. 2001;154(2):267–273. [PMC free article: PMC2150759] [PubMed: 11470816]
- 6.
- Terasawa K, Minami M, Minami Y. Constantly updated knowledge of Hsp90. J Biochem (Tokyo). 2005;137(4):443–447. [PubMed: 15858167]
- 7.
- Beliakoff J, Whitesell L. Hsp90: An emerging target for breast cancer therapy. Anticancer Drugs. 2004;15(7):651–662. [PubMed: 15269596]
- 8.
- Workman P. Altered states: Selectively drugging the Hsp90 cancer chaperone. Trends Mol Med. 2004;10(2):47–51. [PubMed: 15106614]
- 9.
- Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25(1):24–28. [PubMed: 10637609]
- 10.
- Prodromou C, Panaretou B, Chohan S. et al. The ATPase cycle of Hsp90 drives a molecular ‘clamp’ via transient dimerization of the N-terminal domains. EMBO J. 2000;19(16):4383–4392. [PMC free article: PMC302038] [PubMed: 10944121]
- 11.
- Zhao R, Houry WA. Hsp90: A chaperone for protein folding and gene regulation. Biochem Cell Biol. 2005;83(6):703–710. [PubMed: 16333321]
- 12.
- Richter K, Muschler P, Hainzl O. et al. Sti1 is a noncompetitive inhibitor of the Hsp90 ATPase. Binding prevents the N-terminal dimerization reaction during the atpase cycle. J Biol Chem. 2003;278(12):10328–10333. [PubMed: 12525481]
- 13.
- Wegele H, Haslbeck M, Reinstein J. et al. Sti1 is a novel activator of the Ssa proteins. J Biol Chem. 2003;278(28):25970–25976. [PubMed: 12716905]
- 14.
- Panaretou B, Siligardi G, Meyer P. et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol Cell. 2002;10(6):1307–1318. [PubMed: 12504007]
- 15.
- Lotz GP, Lin H, Harst A. et al. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J Biol Chem. 2003;278(19):17228–17235. [PubMed: 12604615]
- 16.
- Wandinger SK, Suhre MH, Wegele H. et al. The phosphatase Ppt1 is a dedicated regulator of the molecular chaperone Hsp90. EMBO J. 2006;25(2):367–376. [PMC free article: PMC1383513] [PubMed: 16407978]
- 17.
- Bali P, Pranpat M, Bradner J. et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: A novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280(29):26729–26734. [PubMed: 15937340]
- 18.
- Kovacs JJ, Murphy PJ, Gaillard S. et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18(5):601–607. [PubMed: 15916966]
- 19.
- Lane CS. Mass spectrometry-based proteomics in the life sciences. Cell Mol Life Sci. 2005;62(7-8):848–869. [PubMed: 15868409]
- 20.
- Zhao R, Davey M, Hsu YC. et al. Navigating the chaperone network: An integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120(5):715–727. [PubMed: 15766533]
- 21.
- Puig O, Caspary F, Rigaut G. et al. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24(3):218–229. [PubMed: 11403571]
- 22.
- Falsone SF, Gesslbauer B, Tirk F. et al. A proteomic snapshot of the human heat shock protein 90 interactome. FEBS Lett. 2005;579(28):6350–6354. [PubMed: 16263121]
- 23.
- Chen B, Piel WH, Gui L. et al. The HSP90 family of genes in the human genome: Insights into their divergence and evolution. Genomics. 2005;86(6):627–637. [PubMed: 16269234]
- 24.
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340(6230):245–246. [PubMed: 2547163]
- 25.
- Passinen S, Valkila J, Manninen T. et al. The C-terminal half of Hsp90 is responsible for its cytoplasmic localization. Eur J Biochem. 2001;268(20):5337–5342. [PubMed: 11606196]
- 26.
- Millson SH, Truman AW, Piper PW. Vectors for N- or C-terminal positioning of the yeast Gal4p DNA binding or activator domains. Biotechniques. 2003;35:60–64. [PubMed: 12866406]
- 27.
- Millson SH, Truman AW, Wolfram F. et al. Investigating the protein-protein interactions of the yeast Hsp90 chaperone system by two-hybrid analysis: Potential uses and limitations of this approach. Cell Stress Chaperones (Winter). 2004;9(4):359–368. [PMC free article: PMC1065275] [PubMed: 15633294]
- 28.
- Millson SH, Truman AW, King V. et al. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p). Eukaryot Cell. 2005;4(5):849–860. [PMC free article: PMC1140089] [PubMed: 15879519]
- 29.
- Uetz P, Giot L, Cagney G. et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403(6770):623–627. [PubMed: 10688190]
- 30.
- Tong AH, Evangelista M, Parsons AB. et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294(5550):2364–2368. [PubMed: 11743205]
- 31.
- Giaever G, Chu AM, Ni L. et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418(6896):387–391. [PubMed: 12140549]
- 32.
- Shoemaker DD, Lashkari DA, Morris D. et al. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat Genet. 1996;14(4):450–456. [PubMed: 8944025]
- 33.
- Winzeler EA, Shoemaker DD, Astromoff A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285(5429):901–906. [PubMed: 10436161]
- 34.
- Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. [PubMed: 16118434]
- 35.
- Nathan DF, Lindquist S. Mutational analysis of Hsp90 function: Interactions with a steroid receptor and a protein kinase. Mol Cell Biol. 1995;15(7):3917–3925. [PMC free article: PMC230631] [PubMed: 7791797]
- 36.
- Wang CW, Stromhaug PE, Shima J. et al. The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J Biol Chem. 2002;277(49):47917–47927. [PMC free article: PMC2754690] [PubMed: 12364329]
- 37.
- Wang CW, Stromhaug PE, Kauffman EJ. et al. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J Cell Biol. 2003;163(5):973–985. [PMC free article: PMC1705953] [PubMed: 14662743]
- 38.
- Dolinski KJ, Cardenas ME, Heitman J. CNS1 encodes an essential p60/Sti1 homolog in Saccharomyces cerevisiae that suppresses cyclophilin 40 mutations and interacts with Hsp90. Mol Cell Biol. 1998;18(12):7344–7352. [PMC free article: PMC109316] [PubMed: 9819421]
- 39.
- Houry WA, Frishman D, Eckerskorn C. et al. Identification of in vivo substrates of the chaperonin GroEL. Nature. 1999;402(6758):147–154. [PubMed: 10647006]
- 40.
- Kerner MJ, Naylor DJ, Ishihama Y. et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122(2):209–220. [PubMed: 16051146]
- 41.
- Iyer K, Burkle L, Auerbach D. et al. Utilizing the split-ubiquitin membrane yeast two-hybrid system to identify protein-protein interactions of integral membrane proteins. Sci STKE. 2005;2005(275):pl3. [PubMed: 15770033]
- 42.
- Miller JP, Lo RS, Ben-Hur A. et al. Large-scale identification of yeast integral membrane protein interactions. Proc Natl Acad Sci USA. 2005;102(34):12123–12128. [PMC free article: PMC1189342] [PubMed: 16093310]
- 43.
- Yan W, Chen SS. Mass spectrometry-based quantitative proteomic profiling. Brief Funct Genomic Proteomic. 2005;4(1):27–38. [PubMed: 15975262]
Publication Details
Author Information and Affiliations
Authors
Rongmin Zhao and Walid A Houry*.Affiliations
Copyright
Publisher
Landes Bioscience, Austin (TX)
NLM Citation
Zhao R, Houry WA. Molecular Interaction Network of the Hsp90 Chaperone System. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.