What Makes PGD for Aneuploidy Screening (AS) by Polar Body (PB) Analysis Interesting and Possible?
Since it has been reported that in humans 90% of embryos are aneuploid as a result of malsegregation mechanisms in maternal meiosis I,1 the detection of abnormal oocytes in IVF treatments has become of considerable importance.
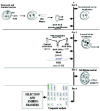
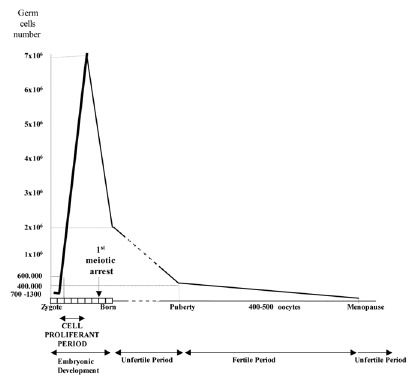
Taking into account the results obtained from the study of recognized pregnancies,2 and gametes,3,4 the conclusion is that the segregation of chromosomes during meiosis is extremely abnormal in humans, and especially during the first meiotic division of the oocytes. This is in agreement with the fact that the first female meiotic division starts during the prenatal stages (at about 11th week of pregnancy) and is not completed until time of ovulation during the fertile life of the woman, between, approximately, 11 and 40 years later2 (fig. 1). Advances in the procedures of assisted human reproduction have facilitated the development of methods of diagnosis which are more precocious than prenatal diagnosis, and corresponds to what is known as Preimplantational Genetic Diagnosis (PGD).5 This has given rise to an availability of biological material (gametes and human embryos), that has provided information for a better estimation of the frequency of aneuploidy in the first stages of embryo development.
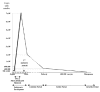
Figure 1
Schematic representation of female gametogenesis.
The results of the application of PGD to aneuploidy screening (AS) are periodically compiled by the European Society of Human Reproduction. The last collection6 includes a total of 2,854 PGD AS cycles performed until december 2001 (71 through the study of the 1PB and 1,783 through the study of blastomeres). Of a total of 24,374 COCs obtained, 20,930 were inseminated and 15,002 were fertilized. A diagnosis was obtained in 9,045 of the 10,760 cells successfully biopsied. There were 3,625 transferable embryos, of which 2,932 were transferred, in 1,342 IVF cycles. A total of 431 biochemical pregnancies were obtained, with 336 pregnancies detected by means of the heartbeat. The success of PGD AS, related to the total number of oocytes, is 18%.
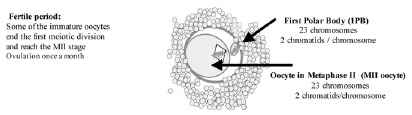
The development of PGD by means of the analysis of the 1PB (PGD-1PB) arose subsequent to PGD on blastomeres.7 The challenge was to detect genetic anomalies in the gametes generated in individuals at risk even before fertilization. This is only feasible if the parent at the risk is the female. The differential characteristics that exist between male and female gametogenesis allow this analysis. While spermatozoa are haploid cells which result from the two meiotic divisions, the oocytes, at the time of ovulation, have not completed meiosis II and are arrested in metaphase of the second meiotic division (fig. 2). The female gamete is made up of two cells, the first of which is called the oocyte in metaphase II (MII oocyte), and the other which is the first polar body (1PB). Both cells contain complementary chromosomal sets. Only the MII oocyte receives the totality of the cytoplasm and has the reproducing function that will be developed upon being fertilized by a sperm. The 1PB does not have any reproductive function and will degenerate. Thus, the 1PB can be biopsied and can be used to carry out an indirect analysis of the genetic contents of the MII oocyte.
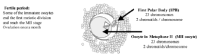
Figure 2
Oocyte at the time of ovulation arrested at Metaphase II.
Methodologies of Analysis of Aneuploidies in Human Oocytes: Cytogenetic Analysis of 1PB and MII of the Oocyte
To carry out a study of aneuploidies of female origin, in a research project, both chromosomal sets should be studied. The result of the 1PB analysis constitutes an internal control of the segregation of the chromosomes, which is essential.
Although one can try to fix both cells at the same time, it is recommended to proceed to the individual fixation of each of them. The cells should be well separated after the total elimination of the zona pellucida with Tyrode's acid or with trypsin solution, or by perforating the zona pellucida, applying diverse alternative methods which allow for obtaining the 1PB, without damaging the MII oocyte.
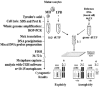
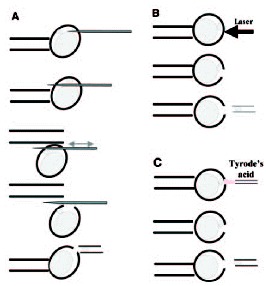
There are diverse biopsy methods of 1PB available, all of which are applied to the mature oocyte, free of cumulus cells, which is placed in an inverted microscope equipped with a micromanipulator, and held with a holding pipette (fig. 3). One of the most common methods is a mechanical system. The oocyte is subjected with its 1PB placed at 12 o'clock, and using a sharp needle pipette in the micromanipulator's arm, a simple or a double slit is made in the zona pellucida.8-10 Then, the needle pipette is removed, and using a biopsy pipette with a diameter of 20-30 μm, the 1PB is gently removed from the oocyte and transferred to a clean, just-filtered drop of medium. The faster system is the laser system that has been used in the mouse.11 The hole is made with just one or two laser shots. The third method, which is the Tyrode's acid system, is not recommended for PGD-1PB because it affects negatively the fertilization capacity of the MII oocyte.12,13
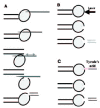
Figure 3
1PB biopsy systems: A) mechanical, B) laser and C) Tyrode's acid.
For the analysis of aneuploidies in oocytes, chromosome spreads of the 1PB and its corresponding MII oocyte fixed on a slide have been mainly used. To obtain adequate chromatin material spread two methods of fixation have been used: the methods of Tarkowsky14 and of Mikamo and Kamiguchi.15
The method of Tarkowsky,15 which is widely used, consists of dropping carnoy fixative solution (methanol: acetic acid (3:1 v/v)) on a previously hypotonized single cell placed on a slide.
In the method of Mikamo and Kamiguchi,15 a gradual fixation of the previously hypotonized oocytes is performed as follows: first, fixative I (methanol: acetic acid: distilled water (5:1:4 v/v, 5 min)); second, fixative II (methanol: acetic acid (3:1 v/v, 10 min)); and third, fixative III (methanol: acetic acid: distilled water (3:3:1 v/v, 1 min)). The quality of the chromosomes is good enough to allow R-banding.16,17
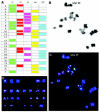
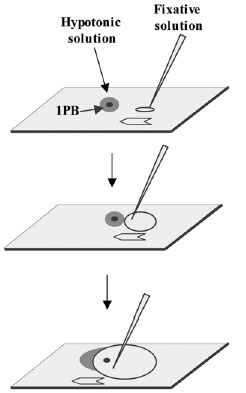
For 1PBs, two fixation methods have been applied: the method of Tarkowsky,14 which is applied in a similar fashion as for the MII oocyte, and the method of Durban.18 This method was developed especially for the fixation of 1PBs and should be performed under stereomicroscopic control. Each previously hypotonized 1PB is fixed on a slide previously treated with trimethoxysylane. Once the 1PB is left on the slide, a soft flow of fixative solution (ethanol: acetic acid; 1:1 v/v) is created which is directed toward the 1PB until it reaches it, dissolving its plasma membrane (fig. 4). This method, applied to 1PBs of fresh oocytes, achieves in 88% of cases, spreads of the chromosomal complement of the 1PB with good chromosomal quality (fig. 5) and has provided the best coincidence between the chromosome complements of the 1PB and the MII oocyte after in situ hybridization techniques (FISH) (fig. 4). In aged oocytes good chromosomal morphology and concordant results are observed with much lower frequency.13
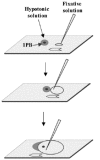
Figure 4
1PB fixation according to Durban's method.

Figure 5
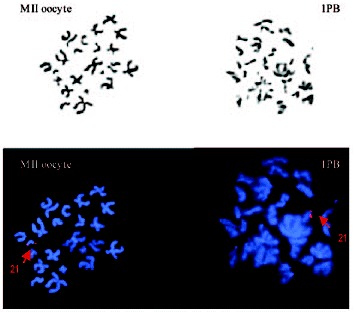
1PB and MII oocyte chromosome complements observed by phase-contrast (up) and by fluorescent microscopy (down). Chromosome 21 is labeled in red.
In general, while the fixed chromosomes of the MII oocyte appear with the two chromatids separated and zig-zagging and with the chromatin quite compact, the chromosomes of the 1PB have their two chromatids also separated, but the chromatin appears more fuzzy and disorganized. Although banding techniques16,17 can be applied to MII chromosomes, it is more recommendable to use FISH techniques with whole-chromosome painting probes, which are noninformative in interphase nuclei, as well as specific probes: locus-specific, centromeric and telomeric among others. FISH has been used for the study of a restricted and predetermined number of chromosomes in one, two or three rounds of FISH. Initially, chromosomes 18 and X19 were analyzed, and subsequently other chromosomes implicated in viable and nonviable aneuploidies in humans were gradually included, eventually reaching, at present, up to nine chromosomes (1, 13, 15, 16, 17, 18, 21, 22 and X).20 These chromosomes have been identified as responsible for aneuploidies in spontaneously abortions and also in blastomeres of embryos of women of advanced age21-23 which in turn would reduce the rate of implantation.19,20,24-26 Treatment of the slide with microwaves for one hour at 75w immediately before hybridization allows reducing the time of FISH and intensifies the fluorescent signals.27
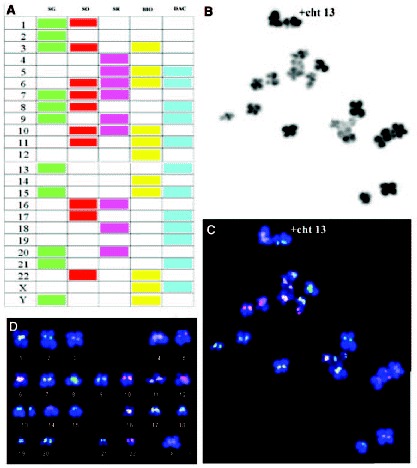
FISH has also been applied to the analysis of the whole chromosomes set4,28,29 by Spectral karyotyping (SKY). The system employs 24 chromosome-specific painting probes. Each probe is labeled with a different proportion of five separate fluorochromes, which on being observed by spectral imaging and with the adequate software, show distinct colors for each chromosome, which permits to correctly identify each chromosome pair in the karyotype. At the same time, a variant of FISH, cenM-FISH, which is a new multicolor-FISH approach applying centromere-specific multicolor-FISH probes directly or indirectly labeled with one, two, three or four distinct fluorochromes,30 has been already applied to the study of meiotic cells.31 This system, when applied to good chromosomal spreads of metaphase II oocytes (fig. 6, unpublished data), appears as an alternative of great interest for the AS analysis in oocytes, because in one single round of FISH all centromeres of the chromosomal complement can be analyzed.
Another technique called the primer in situ (PRINS) has been successfully tested on chromosomal spreads of fixed 1PBs. It is a fast inexpensive method of labeling chromosomes 13, 16, 21 or X, which are frequently involved in aneuploidies.32 Chromosomes are labeled by in situ annealing with chromosome-specific oligonucleotide primers, followed by primer extension with labeled nucleotides using the Taq DNA polymerase. Each 1PB chromosome spread undergoes three successive double-labeling reactions, and it in less than 40 min intense signals are obtained, which is of great interest in the application of PGDs for AS.
But all of the methodologies indicated have a common limitation. They are applied on chromosomal spreads of cells fixed on slides and it is never possible to discard that the hypohaploidies detected are not artifactual losses that occur during fixation.
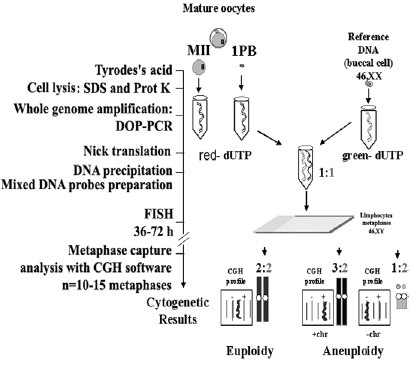
The Comparative Genomic Hybridization (CGH) cytogenetic method applied to isolated cells33,34 is a very favorable alternative for the study of aneuploidies in human oocytes. This method permits one to study all chromosomes of the chromosomal complement of the 1PBs and of the corresponding MII oocyte. The possibility of artifactual losses of chromosomes does not exist, because the cells are introduced, lysed and processed separately inside PCR tubes (fig. 7). This system, given the limitation in the amount of DNA present in an isolated cell (approximately 6pg), requires a preamplification of the whole cell genome using DOP-PCR.35 Next, the preamplified material can be used in CGH. CGH is a competitive DNA hybridization on chromosomal spreads in metaphases of control lymphocytes, with a mixture in equimolecular amounts of the two previously prepared DNAs: DNA of the isolated test cell, labeled with a red fluorochrome (Spectrum red), and a reference DNA, labeled with a green fluorochrome (Spectrum green) coming from an individual with a normal karyotype. The chromosomal losses of the problem cell appear in green, chromosomal gains appear in red and the chromosomal regions that show identical proportions appear in yellow. The limitation of this methodology is that it is not informative for heterochromatic or telomeric regions, and chromosomes 17, 19 and 22 usually show profiles that are difficult to interpret. These limitations are also seen when analyzing biological samples in which there is no limitation in DNA amount.36 Occasionally the results in the 1PB and its corresponding MII oocyte show profiles with losses or gains which do not allow assurance as to whether they affects a whole chromosome or a chromatid.37,38
Study of Aneuploidies in Human Oocytes: 1PB and MII Oocytes
A relevant byproduct of in vitro fertilization techniques has been the availability of discarded oocytes, a fact that has promoted a considerable progress in the knowledge of oocyte and embryo chromosome anomalies and their mechanisms of production. In the best of cases, though sporadically, the oocytes are fresh and have not been inseminated. However, in most cases they are oocytes which are nonfertilized after being inseminated in vitro. In other case, the oocytes are immature. These are aged oocytes that do not represent the oocytes of a general population.
In a recent review,39 the mean frequency of chromosomal alterations seen in the studies that have used the fixation technique of Tarkowsky is 35.98%, which includes 26.5% of aneuploidies (15.5% hypohaploidies, 7.3% hyperhaploidies and 3.6% complex aneuploidies) and 6.02% structural aberrations (deletions, acentric fragments chromosome fragmentation).
No significant differences were observed when cytogenetic results obtained from inseminated oocytes discarded from IVF cycles were compared with those non inseminated or with obtained without hormonal induction.40 No differences observed either when comparing them to those fertilized by ICSI or IVF.38,41
In a recent review,39 a great variability in the incidence of aneuploidies, varying between 2.0% and 59.6%, as well as contradictory results in the effects of maternal age were observed.16,42 In this review the divergences were attributed to the reduced size of the sample of oocytes analyzed and to the difficulty in identifying the chromosomes in MII oocytes, because most studies used only uniform-staining techniques, and in a limited number of studies,43-46 low quality banding techniques.
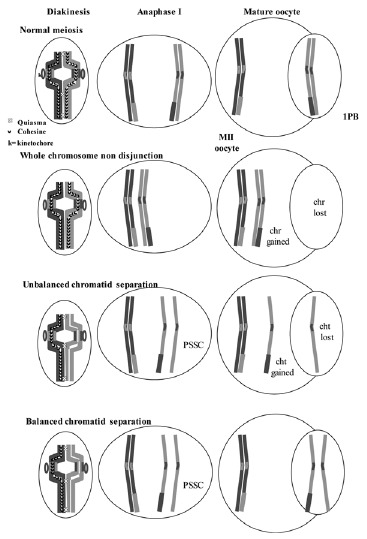
But the uniform-staining techniques on spreads of chromosomal sets of 1PBs and MII oocytes allowed the identification of a new alteration that affected the chromatids of the chromosomes that appeared as a premature separation of sister chromatids (PSSC), which occurred during the first meiotic division.47,48
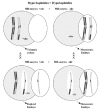
A balanced PSSC is characterized by balanced chromosomal sets with separation of sister chromatids. This anomaly is not considered as responsible for aneuploidies although it may indicate a predisposition to nondisjunction.16,28,49 An unbalanced PSSC, characterized by chromosomal sets with gains or losses of chromatids, is considered as a numerical cytogenetic anomaly, resulting from the early separation and malsegregation of the chromatids of one of the chromosomes of the dyad (fig. 8).

Figure 8
Normal homologous chromosomes and sister chromatid segregation in both meiotic divisions. The mechanisms of non-disjunction, and of unbalanced and balanced sister chromatid separation are shown.
Additionally, in the studies of aneuploidies in oocytes other chromosomal alterations, such as chromosomal loss or gain, produced by non disjunction phenomena (ND) have been described.16,50 While some studies48,51 have observed almost exclusively PSSC, in other cytogenetic studies of oocytes, using FISH with a predetermined number of chromosomes16,20,24,26,28,49 or analyzing all chromosomes simultaneously by CGH,37,38 the presence of both PSSC and ND of chromosomes has been described.
While some works that analyze nonfertilized oocytes and oocytes matured in vitro, suggest that PSSC and ND do not significantly increase over culture time,37,49,52 other studies suggest the opposite.16,53
A study of 1,397 oocytes, fixed by means of the gradual method, indicated that PSSC was significantly increased with respect to ND.54 Similar results have been obtained with different methods of analysis.28,37 At the same time, in fresh, noninseminated oocytes and fresh, unfertilized oocytes from women 35-year old and over, the phenomenon of PSSC is more frequent than ND.28 The authors observed a higher frequency of balanced, rather than unbalanced PSSC in oocytes of women 35 years of age or older, with respect to those of women 24 to 34 years old. This study made it quite clear that balanced PSSC affected more frequently chromosomes of a smaller size.
PSSC may result from premature degradation, before Anaphase I, of the cohesins that maintain sister chromatids joined.55 Some authors have related the presence of PSSC, to advanced maternal age.17,28 Women of advanced age would have an accumulation of mutations in the mitochondrial DNA, not only in the oocytes but also in the follicular cells.56 This fact would reduce mitochondrial function.57 Since the activity of the cohesins depends on ATP,58 a deficiency in oxidative energy metabolism could result in an increase of PSSC in women of advanced age. Also, some authors59,60 have suggested that the presence of free chromatids during the first division could affect the segregation of other chromosomes.
The frequency of aneuploidy varies enormously according to different studies: between 4.7% and 47.5%, with an average of 27.7%, including the studies carried out using R-banding,16,17 FISH techniques with up to 9 chromosomes,19,20,24-26 the SKY system,4,28 or multicolor fluorescent FISH (M-FISH).60 While some authors17,50,61 observe that approximately 16% of the oocytes are aneuploid, the rate of aneuploidy observed in oocytes analyzed by the SKY system4,28 or by the M-FISH system29 varies between 16.7% to 39%, with an average value of 31.3%. At the same time the values obtained by FISH for a limited and predetermined number of chromosomes show even more variation. So, while some groups find frequencies of 3.2%, 9.5% and 4.3% analyzing 2, 6 and 7 chromosomes, respectively (chromosomes 18 and X; 9, 13, 16, 18, 21 and 22 and 1, 9, 13, 16, 18, 21 and X respectively),26,62,63 other groups find higher rates of aneuploidy. Recently, by analyzing 5, 6 or 9 chromosomes (13, 16, 18, 21 and 22; 1, 7, 13, 18, 21 and X, or 1, 13, 15, 16, 17, 18, 21, 22 and X), frequencies of aneuploidies of 41.7%, 44% and 47.5%, have been, respectively, found.20,64,65
The rate of aneuploidy depends on maternal age; thus aneuploidy frequencies of 4%, 9%, 11.5% and 29.8% in the oocytes of females belonging to the maternal age groups of 25-34, 35-39 and 40-45 years old, respectively, had been already estimated.16
In a series of 3,042 nonfertilized oocytes from 792 patients between the ages of 19 and 46 years included in an IVF program, chromosomal alterations which originated by nondisjunction of the chromosomes of the dyad were also correlated to advanced maternal age.17
In spite of the fact that it has been possible to identify that only 25% of the hypohaploidies would correspond to artifacts of the fixation technique or to FISH failures,20 it is still difficult, when only one of the cells is analyzed (1PB or MII oocyte), to know whether the absence of a chromosome should be considered artifactual or not. The rate of aneuploidies is conservatively calculated, in general, as twice the rate of hyperhaploidies.25,26 The rate of aneuploidies observed when applying a combination of methods, i.e., CGH for the 1PBs and FISH for 9 chromosomes for the corresponding MII oocyte, has been estimated at 57.1%.38 This high frequency of aneuploidies could be related to the fact that a greater number of chromosomes are analyzed, and to the fact that the maternal age of these patients was close to 35 years. A very similar frequency, 57.2%, has also been calculated for the 23 chromosomes of the complement from the results obtained from the analysis of 9 chromosomes by FISH.20 Considering the results obtained analyzing 1PBs by CGH37,38 the corresponding MII by FISH in 67 1PB-MII doublets the mean frequency of aneuploidies was 53.7%, with an incidence of 30.5% being in women under 35 years of age and of 69.5% in women 35 or over. The reliability of the study of aneuploidies by FISH variants would be limited by the quality of the extension of the chromosomes and the existence of chromosome overlapping.38 For this reason the use of CGH and FISH for the study of aneuploidies in 1PBs and MII oocytes is a very adequate method for the correct calculation of the rate of aneuploidies.
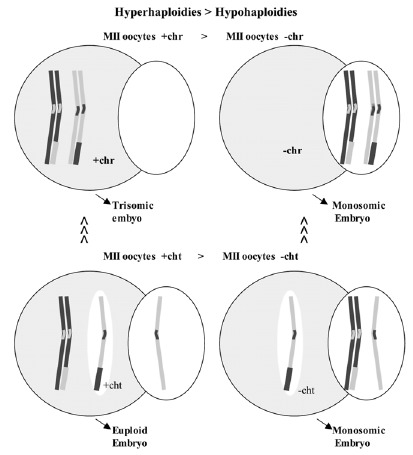
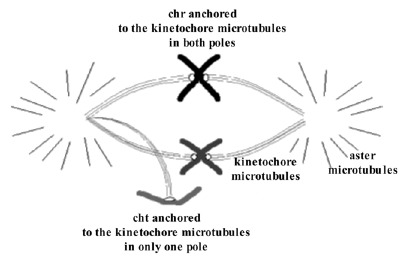
It has been observed that in MII oocytes there is a predominance of hyperhaploidies over hypohaploidies20,64,66 and also a greater proportion of unbalanced PSSC than of chromosome alterations (fig. 9).20,64,66 These results are opposed to those obtained in studies of aneuploidies carried out in wide series of embryos, in which a predominance of monosomies was observed. The difference could be due to losses of chromosomes that could take place in the first mitotic divisions.67 It is not difficult to suppose that the mechanism of chromosomal losses could easily affect free chromatids because they would be weakly joined to one of the poles of the spindle of the second meiotic division (fig. 10). If this were so, the chromatids would tend to be lost in the oocyte cytoplasm. Then, since oocytes with a single-chromatid gain would tend to result in euploid embryos and the oocytes with single-chromatid losses would tend to result in monosomic embryos, the data obtained in MII oocytes and in embryos would not be contradictory (fig. 9).
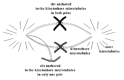
Figure 10
Schematic representation of single-chromatid loss that could easily take place during the second meiotic division due to the fact that the chromatid would be joined to the meiotic spindle in only one of the poles.
Chromosomes belonging to all-chromosome groups have been involved in aneuploidy events. Practically all chromosomes of the human karyotype have been found to be involved in aneuploidies.37,38 The most frequently involved have been chromosomes 1 and 4, followed by chromosomes 22 and X, then by chromosomes 15, 16 and 21, then by chromosomes 2, 3, 6, 7, 8, 9, 13 and 19, and finally by chromosomes 10, 14, 17, 18 and 20. Even though aneuploidies for chromosomes 5, 11 and 12 are not seen in the series, it must be said that on performing a PGD AS analyzing 1PBs by CGH, an aneuploidy affecting chromosome 5 was observed.68 In the same way, in a study combining CGH and SKY, an aneuploidy involving chromosome 12 in an oocyte of a woman carrier of a Robertsonian translocation was found.69
The application of CGH to the study of aneuploidies has allowed for the identification of up to 38.1% more of the aneuploidies that would be identified with FISH for chromosomes 1, 13, 15, 16, 17, 18, 21, 22 and X. Aneuploidies for chromosomes 3, 4, 6, 7, 8, 9, 10, 14 and 19 would have remained unidentified. Despite the fact that oocytes involving multiple aneuploidies would be identified as aneuploid, by FISH, if they were aneuploid for just one of the 9 analyzed chromosomes, 28% of the aneuploid 1PBs, identified by CGH, would have been missed using FISH for the 9 chromosomes.38 These results agree with the 25% of aneuploid blastomeres identified applying CGH that had been erroneously diagnosed with standard FISH.70
Chromosomes 16 and 22 have been frequently involved in aneuploidies in 1PB and in MII oocytes as well as in embryos.20,28,38,64,67 Chromosomes 1, 4 and 17 also show a considerable aneuploidy rate.37 Aneuploidies for chromosome, which are not involved in trisomic offspring or in spontaneous abortions,71 are probably common at the time of conception and would prevent implantation or would induce the death of the implanted embryo.
Chromosome groups E and G are involved in a higher number of aneuploidies due to nondisjunction of the chromosomes of the dyad,48,61,72,73 and also epidemiological results in spontaneous abortions and livebirths show a prevalence of aneuploidies of female origin for the chromosomes of these groups.1,74 Considering these chromosomes of groups A, B and C, including the X chromosome as long chromosomes, and those of groups D, E, F and G as short chromosomes it is seen that short chromosomes are more involved in aneuploidies.26,28,37,38,54 In 1996, errors identified as produced during the second meiotic division were interpreted as derived from anomalies in the first meiotic division, as a result of an increase in the rate of recombination, which would lead to a failure in the separation of the bivalents.75 Currently, it is also believed that a reduction in recombination foci, in the small chromosomes, such as has been shown in human oocytes76 and in human spermatocytes,77 would results in an increased risk of inadequate disjunction of the chromosomes of the dyad and, thus in the production of aneuploid oocytes.65,78,79
The simultaneous study by FISH of both cells, the 1PB and the MII oocyte, in women with a 46XX karyotype indicates the existence of a third mechanism producing aneuploid gametes that originate from trisomic oogonia.20,25 This type of absence of complementarity in studies combining CGH and FISH occurs in approximately 10% of the oocytes studies.37 This fact suggests the existence of gonadal mosaicism in some women who undergo IVF.20,25,26,38 During cell proliferation in the first weeks of embryonic development, some oogonia (fig. 1) would have undergone an anomalous mitotic segregation originating two new distinct cell lines with chromosomal losses and gains, respectively, coexisting with cytogenetically normal lines. This behavior, is individually-dependent and unpredictable, and makes a prenatal diagnosis quite recommendable.
Results of PGD-1PB in AS
In 1990 a PGD variant was proposed, which was initially called Preconception Genetic Diagnosis, and was based on the indirect analysis of oocytes through the genetic study of the 1PB.7
The study of 1PBs is of great interest since, as has been indicated, at least 90% of the chromosomal errors originate in female meiosis. Additionally, this analysis is not affected by the existence of mosaicism, which is very frequent during the first embryonic divisions, and thus makes PGD difficult.80,81 Another of the advantages of this analysis is that in 1PBs, which are arrested in metaphase II, the interpretation of FISH signals is easier than in proliferating cells which can be in any of the stages of the cell cycle.82 This fact can result in errors of interpretation during PGD.81 But the PGD-1PBs has a serious limitation, compared to the study of embryos: they do not provide information on the paternal contribution. This can be solved through the study of one or two blastomeres of the embryo at the 6-8 cell stage.27,83 Neither the biopsy of the 1PBs (and 2PBs) nor that of blastomeres has adverse effects on the development of the embryos.66,83
PGD AS in Older Females
The analysis of aneuploidy is one of the indications of PGD that has grown most and thus has been compiled in the last collection of data of 2005.6 In a recent review,66 the authors indicate that, currently, half of the PGD ASs has been performed through the study of PBs.
Since 1995, a group in Chicago83 applies DGP analyzing simultaneously the 1PB and the 2PB. This allows detecting aneuploidies that originated in both meiotic divisions. The widest series carried out using DGP-PBs by the AS of chromosomes 13, 16, 18, 21 and 22 in women over the age of 35 has shown that more than 50% of the oocytes are aneuploid.64 This series obtained informative FISH results for the chromosomes analyzed in 80% of the cases: in 73.5% of the oocytes in both PBs, in 13.1% only in the 1PB and in 13.4% only in the 2PB. Aneuploidies are observed in 52% of oocytes, and affect more frequently chromatids than chromosomes. The authors observed a comparable rate of errors in meiosis I (41.8%) and meiosis MII (37.3%) in contrast with the traditional concept that aneuploidies mainly originate from female meiosis I. The frequency of errors seen for the chromosomes studied is of 11.68% for chromosome 22, 10.9% for chromosome 21, 6.8% for chromosome 18, 6.4% for chromosome 16 and 6% for chromosome 13. These results do not agree with previous studies85,86,87,88 in which chromosomal aneuploidies were more related to meiosis I or to meiosis II (e.g., for chromosome 18 disomies originated more frequently during the second meiotic division). The authors66 suggest that the errors observed would be more related to anomalies of the meiotic process than to the abnormal segregation of specific chromosomes or chromosome pairs. This could also affect the mitotic process in the embryo.
Results of this group,64 summarizing 1,551 cycles of PGD for 1,027 women of an average age of 38.5 years, from whom a total of 8,293 oocytes were obtained, found that 48% of oocytes are cytogenetically normal for the chromosomes analyzed. In 110 of the IVF cycles a total of 2,587 embryos were transferred and in 21.9% a clinical pregnancy was achieved, finally leading to the birth of 176 healthy babies.
One of the first applications of PGD-1PB for AS was carried out in 10 patients with aneuploidy risk for the offspring due to maternal age and ethical reasons.89 Chromosomes 13, 16, 18, 21 and 22 were analyzed. From the total of 116 mature oocytes available, 93 1PB chromosome complements were obtained three hours after the oocytes were retrieved. FISH-positive results were obtained in 93.5% of the cells. A total of 54 oocytes were euploid and 29 were aneuploid affecting chromosomes 13, 16, 18, 21, and 22 (10.3%, 3.5%, 8.0%, 10.3% and 9.2%, respectively). All euploid oocytes plus 12 undiagnosed oocytes were inseminated 6-7 h after retrieval. From these, 28 embryos were transferred and pregnancy at the heartbeat stage was observed. Three of them suffered spontaneous abortions and one underwent a termination of pregnancy because a 45,X karyotype (Turner Syndrome) was detected by prenatal diagnosis.
In some countries, such as Germany and Switzerland, DGP by analysis of embryo blastomeres is legally forbidden. PGD can only be carried through the study of 1PBs, because this method is considered a preconception diagnosis. Recently, livebirths after PGD with probes for chromosomes 13, 16, 18, 21 and 22 have been reported.90
At the same time, in Germany,91 140 DGP-PBs cycles have been performed in 104 women over the age of 35 using a laser system to biopsy the 1PBs 2-5 h after insemination by ICSI. FISH analysis using probes for chromosomes 13, 16, 18, 21 and 22, on the 1PBs provided results in 88.8% of the oocytes analyzed, of which 50.7% were aneuploid (27.0% disomic, 8.2% nulisomic and 15.5% with multiple anomalies). This high rate of aneuploidy could be related to the advanced age of the patients, and also to the fact that they had a history of implantation failure. Two-hundred-ninety-three of the 443 euploid oocytes were inseminated and fertilized, of which 120 were transferred. Thirty-four biochemical pregnancies produced 33 clinical pregnancies. Five of the pregnancies ended in miscarriages, one of which was a twin pregnancy. Cytogenetic analyses of four of the abortive products showed that they were, respectively, trisomic for chromosome 4, trisomic for chromosome 16 and the other two had a normal karyotype. The pregnancy with trisomy 16 was considered second meiotic origin, although the possibility of a misdiagnosis cannot be excluded. Finally, 24 births were obtained, which gives a rate of “take-home baby” 18.3% per embryo transfer and 15.7% pregnancies per cycle. This means an increase of the rate of clinical pregnancy, the rate of implantation and the rate of “take-home babies” in comparison with the results of the control series without 1PB analysis.
The group of Chicago,92 to increase the number of chromosomes analyzed, pioneered the development of a system that induced the condensation of the chromatin of the 2PB nucleus. The condensation obtained allows the application of G-banding techniques to karyotype the whole chromosome complement. Nevertheless, this methodology is too complex, and has not found a clinical application.
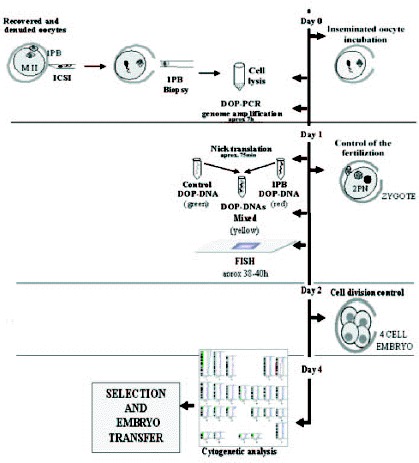
The most groups which perform PGDs-1PB AS analyze a limited number of chromosomes, a fact that represents a serious limitation. The application of the CGH technique to 1PBs of oocytes of a 40-year-old woman68 represented a considerable progress in the evaluation of aneuploidies. The oocytes, once retrieved and denuded, were biopsied and later were fertilized by ICSI.
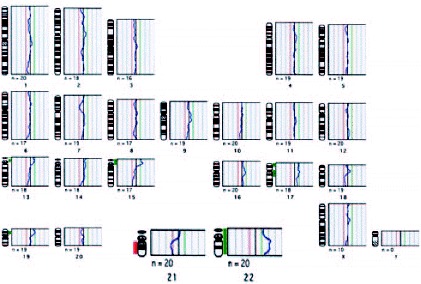
In this study,68 the 1PBs of 11 of the 12 retrieved oocytes were biopsied, and in 10 of them a diagnosis was obtained. Nine of them showed CGH profiles with chromosomal gains or losses for one or two chromosomes. The chromosomes involved were chromosomes 2, 5, 14, 16, 20, 21 and 22, of which four chromosomes (2, 5, 14 and 20) are not usually analyzed by FISH. Only one embryo obtained from a normal oocyte was evolutive, and was transferred on day +3, but no pregnancy was obtained. Aneuploidy was confirmed, by FISH, in the other 9 embryos. Preliminary results from our laboratory on the application of PGD-1PB AS by CGH to a 42-year-old woman, showed a high incidence of aneuploid oocytes. In our protocol, ICSI is carried out within the first two hours after follicular puncture, and immediately after the 1PB is biopsied (fig. 11; unpublished data). Five of the 7 retrieved oocytes were diagnosed, three of them were aneuploid (fig. 12, unpublished data). Three oocytes had alterations for one or two chromosomes, involving chromosomes 19, 21, 22 and X of which one of the chromosomes involved is not analyzed by FISH. Only one of the two cytogenetically normal oocytes detected was evolutive and was transferred on day +4 without achieving a pregnancy.

Figure 12
Aneuploid 1PB chromosome complement diagnosed by CGH showing profiles indicating chromosomal gains of 22 and chromosomal losses of 21. The potential embryo would be trisomic 21 (Down's Syndrome) and monosomic 22.
AS in Female Carriers Balanced Chromosome Rearrangements
In 1963, the possibility that the presence of a chromosomal reorganization could induce alterations in the meiotic segregation of other chromosomes not involved in the reorganization (Interchromosomal effect, ICE) was proposed.93 In recent years, different studies carried out using FISH have shown ICEs in some carriers of chromosomal translocations,94-98 but not in others.95,98,99
When PGD is performed in translocation carriers13,100 and although an acceptable number of normal or balanced embryos can be obtained, the pregnancy rate is disappointingly. PGD by 1PB analysis of chromosomes not involved in the translocation, performed after the analysis of the segregation of the chromosomes involved in translocations (one reciprocal translocation 46XX t(8;13)(q24.1;q22) and two Robertsonian translocations 46XX,t(13;14)(p11;q11)), has been performed.27 The incidence of aneuploidy for chromosomes not involved in the Robertsonian translocations was extremely high (91.7% and 72.7%) and included mainly hyperhaploidies. The frequency of oocytes aneuploid for chromosomes not involved in the rearrangement in carriers of Robertsonian translocations is higher than the maximum value of aneuploidy detected in women with a normal karyotype.37,38,54,81 The reason for this increase could be the tendency to nondisjunction related to advanced maternal age combined with an ICE resulting from the presence of synaptic errors in other chromosome pairs. A high aneuploidy rate of chromosomes not implicated in Robertsonian translocations has already been found by PDG using blastomere analysis.96 It is important to know the chromosome constitution of the oocyte not only for the chromosomes implicated in the reorganization but also those more frequently implicated in aneuploidies21,69 Although the number of theoretically transferred embryos would undoubtedly be much higher if only regarding chromosomes involved in the translocation were analyzed, the pregnancy rate would probably not change and more abortions would probably be produced because aneuploidies could be present in some of the apparently normal or balanced oocytes. The presence of undetected aneuploidies could explain why so many first trimester miscarriages in PGD-1PB cycles were occur after the transfer of apparently normal or balanced embryos. A best approach would be, to include in the analysis of aneuploidies not only women of advanced age but also carriers of reorganizations,96 patients with IVF failures and women who have had spontaneous abortions.
The Future of DGP-1PBs
The application of CGH is a very powerful tool in the diagnosis of aneuploidies that can be applied to the study of 1PBs but which is not applicable to the study of embryo blastomeres at present unless normal embryos are crypreserved to be transferred in the next cycle. The technique requires a very intense dedication of the specialist, or the availability of a good team of molecular cytogeneticists that is well coordinated with a team of embryologists. Recently, methodological advances have been described that can help to make the timing of CGH to be shorter. The use of variants of DOP-PCR-CGH protocols has been proposed applying different Taq polymerases and labeling methods that, more than shortening the procedure allow for the availability of a product of DOP-PCR that is three times greater.101 Additionally, the emerging development of molecular cytogenetic procedures for isolated cells,102,103 along with the implementation of the amplification of multiple displacements (MDA), would permit the detection of aneuploidies using a CGH array.104 A type of CGH array has already permitted the detection of trisomies 13 and 15 in DNA of single cells preamplified with DOP-PCR that should be optimized for the analysis of all chromosomes.105 The Peptide Nucleic Acids (PNAs) can also help to develop DNA chips applicable to the diagnosis of the human chromosomal complement.106 All together, it will soon be possible to carry out PGD in blastomeres without embryos cryopreservation.70 But, in women at risk this should be done simultaneously with the analysis of all of the chromosomal complements of the PBs since that will contribute to a greater understanding of the meiotic chromosomal segregation in women involved in IVF cycles. Moreover, when by law, PGD through blastomere analysis is not possible, PGD through the study of 1PBs, becomes vital since it is the only one in which prefertilization aneuploidy screening is available that avoids embryo biopsy.107
Acknowledgements
This study was supported by Ministerio de Sanidad (FIS PI-020168) and CIRIT (2001 SGR-00201).
References
- 1.
- Nicolaidis P, Petersen M. Origin and mechanisms of no disjunction in human autosomal trisomies. Hum Reprod. 1998;13:313–319. [PubMed: 9557829]
- 2.
- Hassold T, Abruzzo M, Adkins K. et al. Human aneuploidy: Incidence, origin and aetiology. Environ Mol Mutagen. 1996;28:167–175. [PubMed: 8908177]
- 3.
- Shi Q, Martin RH. Aneuploidy in human sperm: A review of the frequency and distribution of aneuploidy, effects of donor age and lifestyle factors. Cytogenet Cell Genet. 2000;90:219–226. [PubMed: 11124518]
- 4.
- Márquez C, Cohen J, Munné S. Chromosome identification in human oocytes and polar bodies by spectral karyotyping. Cytogenet Cell Genet. 1998;81:254–258. [PubMed: 9730613]
- 5.
- Sermon K, von Steirteghem A, Liebaers I. Preimplantation genetic diagnosis. The Lancet. 2004;363:1633–1641. [PubMed: 15145639]
- 6.
- Sermon K, Moutou C, Harper J. et al. ESHRE PGD Consortium data collection IV: May-December 2001. Hum Reprod. 2005;20(1):19–34. [PubMed: 15550497]
- 7.
- Verlinsky Y, Ginsberg N, Liftchez A. et al. Analysis of first polar body: Preconception genetic diagnosis. Hum Reprod. 1990;5:826–829. [PubMed: 2266156]
- 8.
- Malter HE. Partial zona dissection of the human oocyte: A nontraumatic method using micromanipulation to assist zona pellucida penetration. Fertil Steril. 1989;51(1):139–142. [PubMed: 2910709]
- 9.
- Cieslak J, Ivakhnenko V, Wolf G. et al. Three-dimensional partial zona dissection for Preimplantation genetic diagnosis and assisted hatching. Fertil Steril. 1999;71:308–313. [PubMed: 9988403]
- 10.
- Verlinsky Y, Handyside A, Simpson JL. et al. Current progress in Preimplantation genetic diagnosis. J Assist Reprod Genet. 1993;10(5):353–360. [PubMed: 8003878]
- 11.
- Montag M, van der Ven K, Delacretaz G. et al. Laser assisted microdissection of zona pellucida facilitates polar body biopsy. Fertil Steril. 1998;69:539–542. [PubMed: 9531893]
- 12.
- Malter HE, Cohen J. Partial zona dissection of the human oocyte: A nontraumatic method using micromanipulation to assist zona pellucida penetration. Fertil Steril. 1989;51(1):139–148. [PubMed: 2910709]
- 13.
- Durban M, Benet J, Boada M. et al. PGD in carriers of balanced Robertsonian and reciprocal translocations using first polar body analysis. Hum Reprod Update. 2001;7(6):591–602. [PubMed: 11727868]
- 14.
- Tarkowski A. An air drying method for chromosome preparation from mouse eggs. Cytogenetics. 1966;5:394–400.
- 15.
- Mikamo K, Kamiguchi Y. Primary incidences of spontaneous chromosomal anomalies and their origins and causal mechanisms in the Chinese hamster. Mut Res. 1983;108:265–278. [PubMed: 6835223]
- 16.
- Pellestor F. Frequency and distribution of aneuploidy in human female gametes. Hum Genet. 1991;86:283–288. [PubMed: 1997383]
- 17.
- Pellestor F, Andréo A, Arnal F. et al. Maternal ageing and chromosomal abnormalities: New data drown in vitro unfertilized oocytes. Hum Genet. 2003;112:195–203. [PubMed: 12522562]
- 18.
- Durban M, Benet J, Sarquella J. et al. Chromosome studies in first polar bodies from hamster and human oocytes. Hum Reprod. 1998;13(3):583–587. [PubMed: 9572416]
- 19.
- Dailey T, Dale B, Cohen J. et al. Association between nondisjunction and maternal age in meiosis-II human oocytes detected by FISH analysis. Am J Hum Genet. 1996;59:176–184. [PMC free article: PMC1915131] [PubMed: 8659524]
- 20.
- Pujol A, Boiso I, Benet J. et al. Analysis of nine chromosome probes in first polar bodies and metaphase II oocytes for the detection of aneuploidies. Eur J Hum Genet. 2003;11(4):325–336. [PubMed: 12700606]
- 21.
- Bahçe M, Cohen J, Munné S. Preimplantation genetic diagnosis of aneuploidy: Were we looking at the wrong chromosomes? J Assist Reprod Genet. 1999;16:176–181. [PMC free article: PMC3455759] [PubMed: 10224560]
- 22.
- Gianaroli L, Magli MC, Ferraretti AP. et al. Preimplantation diagnosis for aneuploidies in patients undergoing in vitro fertilization with a poor prognosis: Identification of the categories for which it should be proposed. Fertil Steril. 1999;72(5):837–844. [PubMed: 10560987]
- 23.
- Egozcue J, Santaló J, Giménez C. et al. Preimplantation genetic screening and human implantation. J Reprod Immunol. 2002;55:65–72. [PubMed: 12062822]
- 24.
- Verlinsky Y, Cieslak J, Ivakhnenko et al. Prevention of age-related aneuploidies by polar body testing of oocytes. J Assist Reprod and Genet. 1999;16:165–169. [PMC free article: PMC3455760] [PubMed: 10224558]
- 25.
- Mahmood R, Brierley CH, Faed MJW. et al. Mechanisms of maternal aneuploidy: FISH analysis of oocytes and polar bodies in patients undergoing assisted conception. Hum Genet. 2000;106:620–626. [PubMed: 10942110]
- 26.
- Cupisti S, Conn CM, Fragouli E. et al. Sequential FISH analysis of oocytes and polar bodies reveals aneuploidy mechanisms. Prenatal Diagnosis. 2003;23:663–668. [PubMed: 12913873]
- 27.
- Pujol A, Durban M, Benet J. et al. Multiple aneuploidies in the oocytes of balanced translocation: A PGD study using 1PB. Reprod. 2003;126:700–711. [PubMed: 14748689]
- 28.
- Sandalinas M, Márquez C, Munné S. Spectral karyotyping of fresh, noninseminated oocytes. Mol Hum Reprod. 2002;8:580–585. [PubMed: 12029077]
- 29.
- Clyde JM, Hogg JE, Rutherford AJ. et al. Karyotyping of human metaphase II oocytes by multifluor fluorescence in situ hybridization. Fertil Steril. 2003;80:1003–1011. [PubMed: 14556824]
- 30.
- Nietzel A, Rocchi M, Starke H. et al. A new multicolor-FISH approach for characterization of marker chromosomes: Centromere-specific multicolour-FISH (cenM-FISH). Hum Genet. 2001;108:199–204. [PubMed: 11354630]
- 31.
- Oliver-Bonet M, Liehr T, Nietzel A. et al. Karyotyping of human synaptonemal complexes by cenM-FISH. Eur J Hum Genet. 2003;11:879–883. [PubMed: 14571274]
- 32.
- Petit C, Martel-Petit V, Fleurentin A. et al. Use of PRINS for preconception screening of polar bodies for common aneuploidies. Prenat Diagn. 2000;20(13):1067–1071. [PubMed: 11180231]
- 33.
- Wells D, Sherlock JK, Handyside AH. et al. Detailed chromosomal and molecular genetic analysis of single cells by whole genome amplification and comparative genomic hybridisation. Nucleic Acids Research. 1999;27(4):1214–1218. [PMC free article: PMC148305] [PubMed: 9927758]
- 34.
- Voullaire L, Wilton L, Slater H. et al. Detection of aneuploidy in single cells using comparative genomic hybridization. Prenat Diagn. 1999;19:846–851. [PubMed: 10521843]
- 35.
- Telenius H, Carter NP, Bebb CE. et al. Degenerate oligonucleotide-primed PCR: General amplification of target DNA by a single degenerated primer. Genomics. 1992;13:718–725. [PubMed: 1639399]
- 36.
- Moore D, Pallavicini M, Cher M. et al. At statistic for objective interpretation of comparative genomic hybridization (CGH) profiles. Cytometry. 1997;29:183–190. [PubMed: 9222102]
- 37.
- Gutiérrez-Mateo C, Wells D, Benet J. et al. Reliability of comparative genomic hybridization to detect chromosome abnormalities in first polar bodies and metaphase II oocytes. Hum Reprod. 2004;19:2118–2125. [PubMed: 15271867]
- 38.
- Gutiérrez-Mateo C, Benet J, Wells D. et al. Aneuploidy study of human oocytes first polar body comparative genomic hybridization and metaphase II fluorescence in situ hybridization analysis. Huma Reprod. 2004;19(12):2859–2868. [PubMed: 15520023]
- 39.
- Pellestor F, Anahory T, Hamamah S. The chromosomal analysis of human oocytes. An overview of established procedures. Hum Reprod Update. 2005;11(1):15–32. [PubMed: 15569701]
- 40.
- Gras L, McBain J, Trounson A. et al. The incidence of chromosomal aneuploidy in stimulated and unstimulated (natural) inseminated human oocytes. Hum Reprod. 1992;7:1396–1401. [PubMed: 1291565]
- 41.
- Edirisinghe W, Murch A, Junk S. et al. Cytogenetic abnormalities of unfertilized oocytes generated from in vitro-fertilization and intracytoplasmatic sperm injection: A double blind study. Hum Reprod. 1997;12:2784–2791. [PubMed: 9455853]
- 42.
- Zenzes MT, Casper RF. Cytogenetics of human oocytes, zygotes and embryos alter in vitro fertilization. Hum Genet. 1992;88:367–375. [PubMed: 1740312]
- 43.
- Martin RH, Mahadevan MM, Taylor PJ. et al. Chromosomal analysis of unfertile human oocytes. J Reprod Fertil. 1986;78:673–678. [PubMed: 3806521]
- 44.
- Djalali M, Rosenbush B, Wolf M. et al. Cytogenetics of unfertilized human oocytes. J Reprod Fertil. 1988;84:647–652. [PubMed: 3199384]
- 45.
- Pellestor F, Sèle B. Assessment of aneuploidy in the human female by using cytogenetics of IVF failure. Am J Hum Genet. 1988;42:274–283. [PMC free article: PMC1715262] [PubMed: 3341382]
- 46.
- Kola I, Lacham O, Jansen RP. et al. Chromosomal analysis of human oocytes fertilized by microinjection of spermatozoa into the periviteline space. Hum Reprod. 1990;5:575–577. [PubMed: 2394787]
- 47.
- Angell RR. Predivision in human oocytes at meiosis I: A mechanism for trisomy formation in man. Hum Genet. 1991;86:383–387. [PubMed: 1999340]
- 48.
- Angell RR. First-meiotic-division non disjunction in human oocytes. Am J Hum Genet. 1997;61:23–32. [PMC free article: PMC1715867] [PubMed: 9245981]
- 49.
- Munné S, Dailey T, Sultan KM. et al. The use of first polar bodies for preimplantation diagnosis of aneuploidy. Hum Reprod. 1995;10:1014–1120. [PubMed: 7650111]
- 50.
- Zenzes M, Wang P, Casper R. Evidence for maternal predisposition to chromosome aneuploidy in multiple oocytes of some in vitro fertilization patients. Fertil Steril. 1992;57:143–149. [PubMed: 1730308]
- 51.
- Angell RR, Xian J, Keith J. Chromosome abnormalities in human oocytes in relation to age. Hum Reprod. 1993;8:1047–1054. [PubMed: 8408485]
- 52.
- Boiso I, Márquez C, Veiga A. et al. Cytogenetic and fluorescent in situ hybridization analysis of in vitro matured human oocytes. Assist Reprod Rev. 1997;7:160–164.
- 53.
- Pickering S, Johnson M, Braude PR. et al. Cytoeskeletal organization in fresh aged and spontaneously activated human oocytes. Hum Reprod. 1988;3:978–989. [PubMed: 3204153]
- 54.
- Pellestor F, Andréo B, Arnal F. et al. Mechanisms of nondisjunction in human female meiosis: The coexistence of two modes of malsegregation evidenced by the karyotyping of 1397 in-vitro unfertilized oocytes. Hum Reprod. 2002;17:2134–2145. [PubMed: 12151449]
- 55.
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. [PubMed: 9335333]
- 56.
- Bartmann A, Romao G, Ramos E. et al. Why do older women have poor implantation rates? A possible role of the mitochondria. J Ass Reprod Genet. 2004;21:79–83. [PMC free article: PMC3455407] [PubMed: 15202735]
- 57.
- Schon E, Kim S, Ferreira J. et al. Chromosomal nondisjunction in human oocytes: In there a mitochondrial connection? Hum Reprod. 2000;15:160–172. [PubMed: 11041522]
- 58.
- Uhlmann F. The mechanism of sister chromatid cohesion. Exp Cell Res. 2004;296:80–85. [PubMed: 15120997]
- 59.
- Hunt P, LeMaire R, Embury P. et al. Analysis of chromosome behaviour in intact mammalian oocytes: Monitoring the segregation of a univalent chromosome during female meiosis. Hum Mol Genet. 1995;4:2007–2012. [PubMed: 8589675]
- 60.
- Cheng EY, Chen YJ, Bonnet G. et al. An analysis of meiotic pairing in trisomy 21 oocytes using fluorescent in situ hybridization. Cytogenet Cell Genet. 1998;80:48–53. [PubMed: 9678334]
- 61.
- Nakaoka Y, Okamoto E, Miharu N. et al. Chromosome analysis in human oocytes remaining unfertilized after in-vitro insemination: Effect of maternal age and fertilization rate. Hum Reprod. 1998;13:419–424. [PubMed: 9557850]
- 62.
- Dyban A, Freidine M, Severova E. et al. Detection of aneuploidy in human oocytes and corresponding first polar bodies by fluorescent in situ hybridization. J Assist Reprod Genet. 1996;13:73–78. [PubMed: 8825172]
- 63.
- Anahory T, Andréo B, Régnier-Vigouroux G. et al. Sequential multiple probe fluorescence in-situ hybridization analysis of human oocytes and polar bodies by combining centromeric labelling and whole chromosome painting. Mol Hum Reprod. 2003;9:577–585. [PubMed: 12970395]
- 64.
- Kuliev A, Cieslak J, Ilkevitch Y. et al. Chromosomal abnormalities in a series of 6733 human oocytes in preimplantation diagnosis for age-related aneuploidies. Reprod Biomed Online. 2003;6:54–59. [PubMed: 12626143]
- 65.
- Martini E, Flaherty S, Swann N. et al. FISH analysis of six chromosomes in unfertilized human oocytes after polar body removal. J Assist Reprod Genet. 2000;17:276–283. [PMC free article: PMC3455197] [PubMed: 10976415]
- 66.
- Kuliev A, Verlinsky Y. Meiotic and mitotic nondisjunction: Lessons from Preimplantation genetic diagnosis. Hum Reprod Update. 2004;10(5):401–407. [PubMed: 15319376]
- 67.
- Munné S, Bahçe M, Sandalinas M. et al. Differences in chromosome susceptibility to aneuploidy and survival to first trimester. Reprod Biomed Online. 2004;8:81–90. [PubMed: 14759293]
- 68.
- Wells D, Escudero T, Levy B. et al. First clinical application of comparative genomic hybridization and polar body testing for preimplantation genetic diagnosis of aneuploidy. Fertil Steril. 2002;78:543–549. [PubMed: 12215331]
- 69.
- Gutiérrez-Mateo CM, Gadea L, Wells D. et al. Aneuploidy 12 in a Rob (13;14) carrier: Case report Hum Reprod 2005. in press (doi 101093/humrep/deh751) [PubMed: 15695315]
- 70.
- Wilton L, Voullaire L, Sargeant P. et al. Preimplantation aneuploidy screening using comparative genomic hybridization or fluorescence in situ hybridization of embryos from patients with recurrent implantation failure. Fertil Steril. 2003;80:860–868. [PubMed: 14556801]
- 71.
- Simpson J. Incidence and timing of pregnancy losses: Relevance to evaluating safety of prenatal diagnosis. Am J Med Genet. 1990;35:165–173. [PubMed: 2178414]
- 72.
- Kamiguchi Y, Rosenbush CB, Sterzik K. et al. Chromosomal analysis of human oocytes prepared by a gradual fixation air drying method. Hum Genet. 1993;90:533–541. [PubMed: 8428752]
- 73.
- Lim AS, Ho AT, Tsakok MF. Chromosomes of oocytes failing in-vitro fertilization. Hum Reprod. 1995;10:2570–2575. [PubMed: 8567772]
- 74.
- Hassold T, Hunt P. To err (meiotically) is human: The genesis of human aneuploidy. Nat Review Genet. 2001;2(4):280–291. [PubMed: 11283700]
- 75.
- Lamb E, Freeman S, Savage-Austin A. et al. Susceptible chiasmate configurations of chromosome 21 predispose to non disjunction in both maternal meiosis I and meiosis II. Nat Genet. 1996;14:400–405. [PubMed: 8944019]
- 76.
- Tease CH, Hartshorne GM, Hultén MA. Patterns of meiotic recombination in human fetal oocytes. Am J Hum Genet. 2002;70:1469–1479. [PMC free article: PMC379134] [PubMed: 11992253]
- 77.
- Sun F, Oliver-Bonet M, Liehr T. et al. Human male recombination maps for individual chromosomes. Am J Hum Genet. 2004;74:521–531. [PMC free article: PMC1182265] [PubMed: 14973780]
- 78.
- Koehler K, Hawley R, Shreman S. et al. Recombination and non disjunction in humans and flies. Hum Mol Genet. 1996;5(1):1495–1504. [PubMed: 8875256]
- 79.
- Hassold T, Sherman S, Hunt P. Counting cross-overs: Characterizing meiotic recombination in mammals. Hum Mol Genet. 2000;9:2409–2419. [PubMed: 11005796]
- 80.
- Munné S, Cohen J, Weier HUG. Chromosome abnormalities in human arrested preimplantation embryos: A multi-probe FISH study. Am J Hum Genet. 1994;55:150–159. [PMC free article: PMC1918237] [PubMed: 8023843]
- 81.
- Munné S, Cohen J, Sable D. Preimplantation genetic diagnosis for advanced maternal age and other indications. Fertil Steril. 2002;78:234–236. [PubMed: 12137856]
- 82.
- Pujol A, Benet J, Campillo M. et al. The use of a cell-cycle phase-marker may decrease the percentage of errors when using FISH in PGD. Cytogenet Genome Research. 2004;105(1):29–35. [PubMed: 15218255]
- 83.
- Magli MC, Gianaroli L, Ferraretti AP. et al. The combination of polar body and embryo biopsy does not affect embryo viability. Hum Reprod. 2004;19(5):1163–1169. [PubMed: 15070887]
- 84.
- Verlinsky Y, Cieslak J, Freidine M. et al. Pregnancies following preconception diagnosis of common aneuploidies by fluorescent in situ hybridization. Hum Reprod. 1995;10:1923–1927. [PubMed: 8583011]
- 85.
- Sherman SL, Peterson MB, Freeman SB. et al. Non disjunction of chromosome 21 in maternal meiosis I: Evidence for a maternal age dependent mechanism involving reduced recombination. Hum Mol Genet. 1994;3:1529–1535. [PubMed: 7833907]
- 86.
- Hassold T, Merril M, Adkins K. et al. Recombination and maternal age dependent nondisjunction: Molecular studies of trisomy 16. Am J Hum Genet. 1995;57:867–874. [PMC free article: PMC1801507] [PubMed: 7573048]
- 87.
- Peterson MB, Mikkelsen M. Non disjunction in trisomy 21: Origin and mechanisms. Cytogenet Cell Genet. 2000;91:199–203. [PubMed: 11173856]
- 88.
- Fisher JM, Harvey JF, Morton NE. et al. Trisomy 18: Studies of the parent and cell division of origin and effect of aberrant recombination on non disjunction. Am J Hum Genet. 1996;56:669–675. [PMC free article: PMC1801162] [PubMed: 7887421]
- 89.
- Munné S, Sepulveda S, Balmaceda J. et al. Selection of the most common chromosome abnormalities in oocytes prior to ICSI. Prenat Diagn. 2000;20:582–586. [PubMed: 10913958]
- 90.
- Imtburn B, Achermann J, Arter MK. et al. Preimplantation diagnosis in Switzerland—birth of a healthy child alters polar body biopsy. Swiss Med Wkly. 2004;134:259–261. [PubMed: 15243854]
- 91.
- Montag M, van der Ven K, Dorn CH. et al. Outcome of laser-assisted polar body biopsy and aneuploidy testing. Reprod Biomed Online. 2004;9(4):425–429. [PubMed: 15511343]
- 92.
- Verlinsky Y, Evsikov S. Karyotyping of human oocytes by chromosomal analysis of the second polar bodies. Mol Hum Reprod. 1999;5(2):89–95. [PubMed: 10065862]
- 93.
- Lejeune J. Autosomal disordes. Pediatrics. 1963;32:326–337. [PubMed: 14063510]
- 94.
- Blanco J, Egozcue J, Clusellas N. et al. FISH on sperm heads allows the analysis of chromosomal segregation and interchromosomal effects in carriers of structural rearrangements: Results in a translocation carrier, t(5;8)(q33;q13). Cytogenet Cell Genet. 1998;83:275–280. [PubMed: 10072603]
- 95.
- Estop AM, Cieply K, Munné S. et al. Is there an interchromosomal effect in reciprocal translocation carriers? Sperm FISH studies. Hum Genet. 2000;106:517–524. [PubMed: 10914681]
- 96.
- Gianaroli L, Magli MC, Ferraretti AP. et al. Possible interchromosomal effect in embryos generated by gametes from translocation carriers. Hum Reprod. 2002;17(12):3201–3207. [PubMed: 12456624]
- 97.
- Oliver-Bonet M, Navarro J, Carrera J. et al. Aneuploidy and unbalanced sperm in two translocation carriers: Evaluation of the risk. Mol Hum Reprod. 2002;8(10):958–963. [PubMed: 12356948]
- 98.
- Benet J, Oliver-Bonet M, Cifuentes P. et al. Segregation of chromosomes in sperm of reciprocal translocation carriers: A review Cytogenet Genome Res 2005. in press . [PubMed: 16192706]
- 99.
- Honda H, Miharu N, Ohashi Y. et al. Analysis of segregation and aneuploidy in two reciprocal tranlocation carriers, t(3:9)(q26.2;q32) and t(3;9)(p25;q32), by triple-color fluorescence in situ hybridization. Hum Genet. 1999;105:428–436. [PubMed: 10598808]
- 100.
- Munné S, Escudero T, Sandalinas M. et al. Gamete segregation in female carriers of Robertsonian translocation. Cytogenet Cell Genet. 2000;90:303–308. [PubMed: 11124538]
- 101.
- Ozen S, Rechistsky S, Verlinsky Y. Optimization of high-resolution single cell comparative genomic hybridization to fit in PGD framework. Reprod Biomed Online. 2002;4(Supl 2):44.
- 102.
- Wells D. Advances and preimplantation genetic diagnosis. Eur J Obstet Gynecol. 2004;115:s97–s101. [PubMed: 15196725]
- 103.
- Wells D, Steuerwald N, Chu L. et al. Micro array for analysis and diagnosis of human embryosIn: Papp Z, Rodeck C, eds.Recent Advances in Prenatal Genetic DiagnosisBologna, Italy: Medimond,20049–17.
- 104.
- Hellani A, Coskun S, Benkhalifa M. et al. Multiple displacement amplification on single cell and possible PGD application. Mol Hum Reprod. 2004;10(11):847–852. [PubMed: 15465849]
- 105.
- Hu DG, Webb G, Hussey N. Aneuploidy detection in single cells using DNA array-based comparative genòmic hybridisation. Mol Hum Reprod. 2004;10:283–289. [PubMed: 14985479]
- 106.
- Pellestor F, Paulasova P. The peptide nucleic acids, efficient tools for molecular diagnosis. Int J Mol Med. 2004;13(4):521–525. [PubMed: 15010851]
- 107.
- Gitlin SA, Gibbons WE, Gosden RG. Oocyte biology and genetics revelations from polar bodies. Reprod Biomed Online. 2003;6(4):403–309. [PubMed: 12831582]
Publication Details
Author Information and Affiliations
Authors
J Navarro,* C. Gutiérrez-Mateo, A. Pujol, M. Durban, J.F. Sánchez-García, J. Egozcue, and J. Benet.Affiliations
Copyright
Publisher
Landes Bioscience, Austin (TX)
NLM Citation
Navarro J, Gutiérrez-Mateo C, Pujol A, et al. Preimplantation Genetic Diagnosis (PGD): Screening for Aneuploidy in Human Oocytes and Polar Bodies. In: Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.