NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Nonsense-mediated mRNA decay (NMD) in higher eukaryotes requires the phosphorylation and dephosphorylation of Upf1, an RNA helicase. Recent studies suggest that sequential phosphorylation and dephosphorylation of Upf1 follows the recognition of premature termination codons within mRNAs and is required for NMD. Upf1 phosphorylation, which is mediated by the phosphoinositide 3-kinase-related protein kinase SMG-1, triggers remodeling of the Upf1-containing surveillance complex to form another complex. This second complex, which contains SMG-5, SMG-7 and protein phosphatase 2A, is required for Upf1 dephosphorylation. Upf1 phosphorylation requires a SMG-1-Upf1 complex that interacts directly with the exon junction complex component Upf2, supporting the conclusion that Upf1 phosphorylation occurs on mRNA during the mRNA surveillance process.
Introduction
NMD is a surveillance mechanism that detects and degrades mRNAs containing premature termination codons (PTCs). Seven smg gene products have been identified as trans-acting factors that are required for NMD in Caenorhabditis elegans (Table 1; see chapter by Anderson), and each is conserved in mammals (Table 1; see chapters by Maquat, and Singh and Lykke-Andersen). One of these mammalian factors is Upf1 (SMG-2 in C. elegans), which undergoes a cycle of phosphorylation and dephosphorylation that is required for NMD. The other mammalian smg gene products can be divided into two groups based on function (Table 1). One group consists of SMG-1, Upf2 (SMG-3 in C. elegans) and Upf3 (SMG-4 in C. elegans), which are needed to phosphorylate Upf1. The other group consists of SMG-5, SMG-6, and SMG-7, which are required to dephosphorylate Upf1. Upf1 forms a surveillance complex with other Upf/SMG proteins. Other lines of evidence suggest that an exon junction complex (EJC) helps distinguish PTCs from normal translation termination codons in vertebrate cells. However, the biochemical nature of the surveillance complex, as well as the biological significance of the interactions between the surveillance complex and the EJC, are not well understood. Recent studies that aimed to clarify the roles of Upf1 phosphorylation and dephosphorylation during NMD and the interactions among proteins required for NMD not only establish the importance of both Upf1 phosphorylation and dephosphorylation but also reveal the mechanism by which the surveillance complex recognizes a PTC within mRNA.
Table 1
Evolutionary relationships of Upf/SMG proteins.
Structure and Biochemical Characterization of SMG-1
SMG-1 is the fifth member of a family of phosphoinositide 3-kinase-related protein kinases (PIKKs) that includes ATM (ataxia telangiectasia mutated), ATR (ATM and Rad3 related), mTOR (target of rapamycin) and DNA-PK (DNA-dependent protein kinase)1 (see chapter by Abraham et al). Human and C. elegans SMG-1 share the highest overall similarity among PIKKs. SMG-1 shares PIKK and helical repeat regions (containing FAT domains) with all the PIKK family members (Fig. 1). SMG-1 activity shows preference for Mn2+ over Mg2+, is inhibited in vitro by wortmannin (IC50 of 60-105 nM) and caffeine (IC50 of 0.3 mM), but is not inhibited in vitro by staurosporin nor rapamycin.2-4 Wortmannin inhibits SMG-1 with an IC50 value of 1-2 μM in intact cells.3,4 ATM, ATR and DNA-PK are S/T-Q-directed kinases,1 and they show strong preference for phosphorylating serine (S)/threonine (T) followed by a glutamine (Q). Similarly to these PIKKs, SMG-1 preferentially phosphorylates S/T-Q, and it can phosphorylate p53 at serine 15.3,4 SMG-1 and mTOR contain a TS (TOR, SMG-1 homology) domain, which consists of a binding site for the FKBP12-rapamycin complex in mTOR. However, there is no evidence suggesting that SMG-1 interacts with the FKBP12-rapamycin complex. The FKBP12-rapamycin-binding (FRB) domain essential for mTOR activation has been reported to bind phosphatidic acid (PA).5 However, again, there is no evidence that SMG-1 interacts with PA.
In addition to the sequences conserved among all PIKK family members, there are SMG-1-specific sequences that may be involved in SMG-1-specific protein-protein interactions. Consistent with this notion, there are proteins that bind specifically to the N-terminal half of SMG-1 (Kashima et al, in preparation). Another unique structural feature of SMG-1 relative to other PIKKs is a large insert between the PIKK domain and the FAT-C domain. Although we do not know the role of this insert, its presence within the SMG-1 proteins of all species examined to date implies functional significance.
Human SMG-1 has at least two isoforms, p430 and p400, which have different N-terminal sequences (see also chapter by Abraham and Oliveira). An antibody against the N-terminal 106 amino acids of the 3657 amino acid-protein encoded by cDNA recognizes only the p430 isoform.3 The p400 isoform lacks antibody recognition. The biological significance of this N-terminal sequence variation remains unclear.
SMG-1-Mediated Phosphorylation of Upf1
While genetic screens of C. elegans identified the seven smg genes required for NMD, genetic screens of Saccharomyces cerevisiae identified only the three upf genes required for NMD (Table 1; see also chapter by Baker and Parker). Therefore only the Upf1/SMG-2, Upf2/SMG-3 and Upf3/SMG-4 core components of the NMD surveillance complex are conserved in both eukaryotes. Knockdown of each Drosophila melanogaster6 or human Upf/SMG protein suppresses NMD.4,6-8 Additionally, components of the vertebrate exon junction complex (EJC) in addition to the Upf proteins have also been shown to be required for NMD (see chapter by Maquat). C. elegans SMG-2 is a phosphoprotein, and genetic studies have classified the six other smg proteins as regulators of SMG-2 phosphorylation.9 SMG-1, SMG-3 and SMG-4 are required for SMG-2 phosphorylation, whereas SMG-5, SMG-6 and SMG-7 are required for SMG-2 dephosphorylation.9
Similar to C. elegans SMG-2, mammalian Upf1 is a phosphoprotein,3,10 and SMG-1 kinase activity is essential for Upf1/SMG-2 phosphorylation and NMD in vivo.3,11 SMG-1 phosphorylates Upf1/SMG-2 in vitro, suggesting that SMG-1 is a direct kinase of Upf1/SMG-2.2,3,11 SMG-1 phosphorylates N-terminal and C-terminal regions of Upf1 in vitro.3 Four serine residues, LS1073QP, LS1078QD, LS1096QD and LS1116QY, in the metazoan-specific C-terminal SQ-rich region of human Upf1 are efficiently phosphorylated by SMG-1 in vitro.3 At least two of these residues, S1078 and S1096, are also phosphorylated in vivo in an SMG-1-dependent manner.3 The sequence motifs surrounding S1078 and S1096 are conserved among metazoan Upf1s, supporting the importance of phosphorylation at these sites (Fig. 2). Furthermore, expression of phospho-resistant Upf1, in which the four serine residues targets of SMG-1-mediated phosphorylation were replaced with alanine, suppresses NMD in mammalian cells (Kashima et al, in preparation). All these results strongly suggest that NMD involves Upf1 phosphorylation at the four sites.
Only a very small fraction of Upf1 is phosphorylated in mammalian cells.3,10,12 Importantly, overexpression of SMG-1 induces accumulation of highly phosphorylated Upf1 and accelerates NMD in mammalian cells.3 Thus, SMG-1-mediated phosphorylation of Upf1 seems to be a critical rate-limiting step during NMD.
Mechanism of SMG-1-Mediated Phosphorylation of Upf1
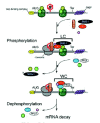
SMG-3/Upf2 is essential for the SMG-1-mediated phosphorylation of Upf1 in both C. elegans and mammals9 (Kashima et al, in preparation). Interestingly, SMG-1 associates with SMG-2 in a SMG-3-independent manner in C. elegans,11 and human SMG-1 associates with Upf1 in a Upf2-independent manner (Kashima et al, in preparation). However, even though SMG-1 forms a complex with SMG-2, SMG-1 fails to phosphorylate SMG-2 in a smg-3 C. elegans.9,11 A very similar situation applies to mammalian-cell counterparts (Kashima et al, in preparation). Additionally, mutated Upf1 that cannot associate with Upf2 is scarcely phosphorylated by SMG-1, although its association with SMG-1 remains intact in mammalian cells. These observations led us to hypothesize a model for Upf1 phosphorylation during NMD (Fig. 3). In the model, prior to phosphorylation by SMG-1, Upf1 associates with SMG-1 to form an initial SMG-1-Upf1 complex (IC). Subsequently, Upf2 associates with the IC and triggers Upf1 phosphorylation.
Upf1 can associate with eukaryotic translation release factor (eRF) 1 and 3 in yeast13 and mammalian cells (Y.K. Kim and L.E. Maquat, unpublished data; G. Singh and J. Lykke-Andersen, unpublished data; I. Kashima and S. Ohno, unpublished data), and Upf2 is most likely an EJC component (see chapters by Maquat and Singh and Lykke-Andersen). Taken together, the finding that SMG-1-mediated phosphorylation of SMG-2/Upf1 requires SMG-3/Upf2 in C. elegans9 and mammals (Kashima et al, in preparation) suggests that SMG-1-mediated Upf1 phosphorylation occurs on EJCs after PTC recognition (Fig. 3). This notion is supported by the following observations. First, the phosphorylated form of Upf1 is primarily present in the polysomal fraction of mammalian cells.10 Second, SMG-1 immunoprecipitates with EJC components in a Upf2-dependent manner (Kashima et al, in preparation). Third, SMG-1 immunoprecipitates with poly(A) binding protein (PABP) and cap binding protein (CBP) 20 only when cell extracts are not treated with RNase (Kashima et al, in preparation).
Consequences of SMG-1-Mediated Phosphorylation of Upf1
To date, the ATPase activities of nonphosphorylated and phosphorylated Upf1 are not detectably different (I. Kashima and T. Ohnishi, unpublished data). On the other hand, there is evidence that phosphorylation induces remodeling of the Upf1-containing surveillance complex. SMG-5 and SMG-7 interact and associate with phosphorylated (P)-Upf1 to form the wee complex (WC)12,14 (Fig. 3). WC contains the catalytic subunit of protein phosphatase 2A (PP2A) and Upf3aS.12 As noted above, SMG-5 and SMG-7 are essential for Upf1/SMG-2 dephosphorylation12,14 (Kashima et al, in preparation).
Although sequence analysis does not predict SMG-5 and SMG-7 to be either phosphatases or kinases, they appear to be targeting subunits of PP2A. Okadaic acid, a potent inhibitor of PP2A, increases accumulation of P-Upf1 and the presence of WC in vivo.3,12 Thus, PP2A is most likely a P-Upf1 phosphatase. The N-terminal sequence of SMG-7, including the TPR (tetratricopeptide repeat) motif, shows structural similarity to 14-3-315 and associates with the SQ-rich sequence within the Upf1 C-terminus in a phospho-specific manner.15 Consistently, a phosphorylation-resistant variant of Upf1 in which four serine residues that are normally targeted for phosphorylation by SMG-1 are replaced by alanine shows reduced affinity for the SMG-5-SMG-7 complex (Kashima et al, in preparation). The 14-3-3-like domains are conserved in SMG-5, SMG-6 and SMG-7, suggesting that they can also bind the phosphorylated Upf1 peptide sequence (Fig. 1C). Additionally, SMG-5 and SMG-6 share PIN (PilT amino-terminal) and FS-C (SMG-5 [Five]-SMG-6 [Six] C-terminus) domains within their C-termini (Fig. 1C). A PIN domain is found in many proteins having nuclease activity16 (Fig. 1C). SMG-5 harboring an asparatic acid (D)-860 to alanine (A) substitution with the PIN domain or a deletion that removes the FS-C domain maintain the ability to selectively bind P-Upf1, Upf3aS, SMG-7 and PP2A but fail to support P-Upf1 dephosphorylation and, when overexpressed, block NMD. Thus, both PIN and FS-C domain are essential for SMG-5 function in P-Upf1 dephosphorylation and NMD. D860 within the PIN domain is strictly conserved within SMG-5 and SMG-6 as well as PIN domains from other proteins and is thought to be essential for catalytic activity16 (Fig. 1C). Therefore, it is possible that SMG-5 and SMG-6 have nucleotide binding and/or nuclease activity, which might be essential to trigger dephosphorylation of P-Upf1 and NMD. SMG-6 has been shown to associate with Upf1, Upf2, Upf3b and PP2A, although it is unclear whether it also associates with P-Upf1, SMG-5 and SMG-7.17 To date, we do not know the role of SMG-6 on the mRNA surveillance complex.
The SMG-1-independent interaction between SMG-2 and SMG-5 in C. elegans, and the association of recombinant, nonphosphorylated amino acids 1-462 of human Upf1 with human SMG-5 in vitro suggest that SMG-5, and probably SMG-7, can also associate with Upf1/SMG-2 in a phosphorylation-independent manner.12,14 Upf1 harboring a truncated metazoan-specific, N-terminal region (NCR, N-terminal conserved region, see Fig. 2B) has a reduced affinity for the SMG-5-SMG-7 complex and is more highly phosphorylated than wild-type Upf1 in vivo.12 The above observations not only support the importance of the SMG-5-SMG-7 complex for P-Upf1 dephosphorylation, but also suggest that the NCR is a primary binding domain of the SMG-5-SMG-7 complex. Initially, the SMG-5-SMG-7 complex associates with the NCR, possibly in a phosphorylation-independent manner, and then binds the phosphorylated SQ-motif within the C-terminal region of Upf1. How can the SMG-5-SMG-7 complex support dephosphorylation of Upf1 by PP2A? One possibility is that the SMG-5-SMG-7 complex supports the action of PP2A in WC. Another possibility is that, similar to the situation for IC, WC needs additional factor(s) to trigger Upf1 dephosphorylation, and the SMG-5-SMG-7 complex recruits the activator(s) of PP2A. At present, we cannot determine which, if either, of these possibilities is applicable.
Upf2 makes a large, ˜1000-kDa protein complex (LC) that contains Upf1 and Upf3aL18 but not SMG-5, SMG-7, Upf3aS12,18 (Fig. 3). Intriguingly, Upf1 harboring an NCR deletion associates with both Upf2 and Upf3aL more strongly than does wild type Upf1, suggesting that the association of the SMG-5-SMG-7 complex with P-Upf1 results in release of Up2, Upf3aL and possibly SMG-1. WC complex formation then induces the dephosphorylation of P-Upf1. We do not know much about SMG-5-SMG-7 complex function except in P-Upf1 dephosphorylation. Yellow fluorescent protein-tagged SMG-7 recruits Cyan fluorescent protein-tagged Upf1 to cytoplasmic foci (known as P-bodies or GW-bodies) (see chapters by Baker and Parker, and Singh and Lykke-Anderson),19 suggesting that SMG-7 might target mRNA to P-bodies. The specific association of P-Upf1 and SMG-7 suggests that Upf1 dephosphorylation might occur in P-bodies. Consistent with this possibility, artificially tethering SMG-7 to mRNA results in mRNA decay independently of an upstream termination codon.19 The role of P-Upf1 dephosphorylation might be to trigger the mRNA degradation machinery.
Regulation of SMG-1-Mediated Upf1 Phosphorylation
The amount of P-Upf1 in mammalian cells is reduced by serum starvation.10 Additionally, NMD is inhibited by the amino acid starvation.20 Serum and amino acid starvation suppresses translation.21,22 Therefore, inhibition of Upf1 phosphorylation and NMD by starvation may be simply because of reduced translation. Serum and amino acid starvation suppress the mTOR signaling cascade, which positively regulates translation.1 On the other hand, rapamycin, which is a specific inhibitor of mTOR, does not inhibit NMD or Upf1 phosphorylation in mammalian cells.3 Overproduction of 4E-binding protein (BP)1, which is regulated by mTOR, cannot suppress NMD, since it inhibits eukaryotic translation initiation factor (eIF)4E-dependent but not CBP80/CBP20-dependent translation23 (see chapter by Maquat). Taken together, these observations suggest that SMG-1 might respond to extracellular stimulation and be inactivated by serum and/or amino acid starvation. Indeed, SMG-1 is a stress-activated kinase that is stimulated by ionizing radiation (IR) and UV-B.4 Genotoxic stress induces SMG-1- and ATM-dependent Upf1 phosphorylation in mammalian cells4 It will be interesting to define the role of Upf1 phosphorylation after genotoxic stress. Unlike in C. elegans, SMG-1-deficiency in adult D. melanogaster does not abolish NMD.24 In this situation, ATM might regulate Upf1 phosphorylation, although no significant role of ATM in NMD has been identified in mammalian cells. SMG-1 can also contribute to the regulation of p53 activation (see chapter by Abraham and Oliveira).
Suppression of SMG-1-Mediated Upf1 Phosphorylation Inhibits NMD
The finding that NMD requires Upf1 phosphorylation and dephosphorylation raises the promise of experimental strategies that specifically suppress NMD in mammalian cells. Inhibitors of SMG-1, the overproduction of kinase-inactive SMG-1 or SMG-1 knockdown can inhibit both Upf1 phosphorylation and NMD in mammalian cells3,4,25 (Usuki et al, in preparation). These inhibitors also prevent the decay of endogenous nonsense-containing p53 mRNA in two cancer cells3 and nonsense-containing collagen VI α2 mRNA in Ullrich's disease fibroblasts.25 The inhibition of SMG-1 results in accumulation of C-terminus truncated proteins in vivo.3,25 Those results were confirmed by SMG-1 knockdown4 (Usuki et al, in preparation). Notably, in case of Ullrich's disease fibroblasts, up-regulation of a truncated collagen VI α2 subunit results in collagen VI assembly and partially functional extracellular matrix formation25 (Usuki et al, in preparation). NMD protects cells by preventing aberrant truncated proteins that manifest gain-of-function and dominant-negative effects that can result in genetic diseases (see chapter by Holbrook et al). In some cases, such as Ullrich's fibroblasts, truncated protein retains normal functions at least partially, and the selective inhibition of NMD may provide a novel strategy to rescue the disease phenotype in this and other PTC-related diseases where the truncated protein shows no dominant-negative effect. However, pharmacological approaches using wortmannin or caffeine, which is another inhibitor of SMG-1, cannot be administered to patients because of toxicity and nonspecific side effects. Identification of specific inhibitors of SMG-1 needs to be established for useful clinical applications.
Anyway, suppression or inhibition of Upf1 phosphorylation has provided a rather selective suppression of NMD under a variety of experimental conditions. This has greatly not only clarified the physiological role of NMD in mammalian cells but also identified target genes regulated by NMD in a variety of physiological and pathological conditions.
Perspectives
As described above, accumulating evidence suggests that Upf1 phosphorylation by SMG-1 occurs upon PTC recognition during the mRNA surveillance process. Recently, phosphorylation of Upf1 protein (p) was also described in yeast.26 As in metazoans, phosphorylated Upf1p in yeast is less abundant than unphosphorylated Upf1p. Yeast Upf1p does not contain the C-terminal SQ-rich and NCR regions of Upf1/SMG-2 that mediate SMG-5-SMG-7 complex binding (Fig. 2). One intriguing possibility is that Tor1p and Tor2p are Upf1p kinases. Tor1p and Tor2p are most closely related to the SMG-1 PIKK family member (Fig. 1A), but they do not preferentially target the S/T-Q motif.1 While no obvious orthologs to SMG-1, SMG-5, SMG-6 and SMG-7 are present in the yeast genome, there are four proteins, Nmd4p, Yor166cp, Est1p and EBS1p, that resemble SMG-5, SMG-6 and SMG-7 that could function like metazoan SMG-5, SMG-6 and SMG-7. For instance, Nmd4p and Yor166cp contain a PIN domain related to that of SMG-5 and SMG-616 (Fig. 1C), and Nmd4p interacts with Upf1p.27 Est1p, which has been identified as a putative homolog to SMG-6 (see chapter by Azzalin et al), and Ebs1p, both of which are essential for telomerase function, have a 14-3-3 like domain, although they lack several phosphoserine binding residues.15 It remains to be clarified whether the cycle of Upf1 phosphorylation and dephosphorylation is critical for NMD in yeast as it is in metazoans.
Acknowledgements
We thank L. Maquat and J. Lykke-Andersen for communicating unpublished results at the time of writing. This work was supported in part by grants to S. Ohno from the Japan Society for the Promotion of Science, from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and from the Collaboration of Regional Entities for the Advancement of Technological Excellence program of Japan Science and Technology Agency. A. Yamashita received support from the Yokohama Foundation for Advancement of Medical Science. I. Kashima is a Research Fellow of the Japan Society for the Promotion of Science.
References
- 1.
- Abraham RT. PI 3-kinase related kinases: ‘Big’ players in stress-induced signaling pathways. DNA Repair (Amst). 2004;3(89):883–887. [PubMed: 15279773]
- 2.
- Denning G, Jamieson L, Maquat LE. et al. Cloning of a novel phosphatidylinositol kinase-related kinase: Characterization of the human SMG-1 RNA surveillance protein. J Biol Chem. 2001;276(25):22709–22714. [PubMed: 11331269]
- 3.
- Yamashita A, Ohnishi T, Kashima I. et al. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15(17):2215–2228. [PMC free article: PMC312771] [PubMed: 11544179]
- 4.
- Brumbaugh KM, Otterness DM, Geisen C. et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14(5):585–598. [PubMed: 15175154]
- 5.
- Fang Y, Vilella-Bach M, Bachmann R. et al. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294(5548):1942–1945. [PubMed: 11729323]
- 6.
- Gatfield D, Unterholzner L, Ciccarelli FD. et al. Nonsense-mediated mRNA decay in Drosophila: At the intersection of the yeast and mammalian pathways. EMBO J. 2003;22(15):3960–3970. [PMC free article: PMC169044] [PubMed: 12881430]
- 7.
- Mendell JT, ap Rhys CM, Dietz HC. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298(5592):419–422. [PubMed: 12228722]
- 8.
- Kim YK, Furic L, Desgroseillers L. et al. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell. 2005;120(2):195–208. [PubMed: 15680326]
- 9.
- Page MF, Carr B, Anders KR. et al. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19(9):5943–5951. [PMC free article: PMC84455] [PubMed: 10454541]
- 10.
- Pal M, Ishigaki Y, Nagy E. et al. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA. 2001;7(1):5–15. [PMC free article: PMC1370068] [PubMed: 11214180]
- 11.
- Grimson A, O'Connor S, Newman CL. et al. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans. Mol Cell Biol. 2004;24(17):7483–7490. [PMC free article: PMC506987] [PubMed: 15314158]
- 12.
- Ohnishi T, Yamashita A, Kashima I. et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12(5):1187–1200. [PubMed: 14636577]
- 13.
- Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV. et al. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12(11):1665–1677. [PMC free article: PMC316864] [PubMed: 9620853]
- 14.
- Anders KR, Grimson A, Anderson P. SMG-5, required for C.elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 2003;22(3):641–550. [PMC free article: PMC140740] [PubMed: 12554664]
- 15.
- Fukuhara N, Ebert J, Unterholzner L. et al. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17(4):537–547. [PubMed: 15721257]
- 16.
- Clissold PM, Ponting CP. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr Biol. 2000;10(24):R888–890. [PubMed: 11137022]
- 17.
- Chiu SY, Serin G, Ohara O. et al. Characterization of human Smg5/7a: A protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA. 2003;9(1):77–87. [PMC free article: PMC1370372] [PubMed: 12554878]
- 18.
- Schell T, Kocher T, Wilm M. et al. Complexes between the nonsense-mediated mRNA decay pathway factor human upf1 (up-frameshift protein 1) and essential nonsense-mediated mRNA decay factors in HeLa cells. Biochem J. 2003;373(Pt 3):775–783. [PMC free article: PMC1223536] [PubMed: 12723973]
- 19.
- Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16(4):587–596. [PubMed: 15546618]
- 20.
- Mendell JT, Sharifi NA, Meyers JL. et al. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36(10):1073–1078. [PubMed: 15448691]
- 21.
- Pain VM. Translational control during amino acid starvation. Biochimie. 1994;76(8):718–728. [PubMed: 7893822]
- 22.
- Duncan R, Hershey JW. Regulation of initiation factors during translational repression caused by serum depletion. Abundance, synthesis, and turnover rates. J Biol Chem. 1985;260(9):5486–5492. [PubMed: 3988764]
- 23.
- Chiu SY, Lejeune F, Ranganathan AC. et al. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18(7):745–754. [PMC free article: PMC387415] [PubMed: 15059963]
- 24.
- Chen Z, Smith KR, Batterham P. et al. Smg1 nonsense mutations do not abolish nonsense-mediated mRNA decay in Drosophila melanogaster Genetics 2005 . In press. [PMC free article: PMC1456532] [PubMed: 15965240]
- 25.
- Usuki F, Yamashita A, Higuchi I. et al. Inhibition of nonsense-mediated mRNA decay rescues the phenotype in Ullrich's disease. Ann Neurol. 2004;55(5):740–744. [PubMed: 15122717]
- 26.
- de Pinto B, Lippolis R, Castaldo R. et al. Overexpression of Upf1p compensates for mitochondrial splicing deficiency independently of its role in mRNA surveillance. Mol Microbiol. 2004;51(4):1129–1142. [PubMed: 14763985]
- 27.
- He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9(4):437–454. [PubMed: 7883168]
- Introduction
- Structure and Biochemical Characterization of SMG-1
- SMG-1-Mediated Phosphorylation of Upf1
- Mechanism of SMG-1-Mediated Phosphorylation of Upf1
- Consequences of SMG-1-Mediated Phosphorylation of Upf1
- Regulation of SMG-1-Mediated Upf1 Phosphorylation
- Suppression of SMG-1-Mediated Upf1 Phosphorylation Inhibits NMD
- Perspectives
- Acknowledgements
- References
- Role of SMG-1-mediated Phosphorylation of Upf1 in NMD - Madame Curie Bioscience ...Role of SMG-1-mediated Phosphorylation of Upf1 in NMD - Madame Curie Bioscience Database
- Centrins, a Novel Group of Ca2+-Binding Proteins in Vertebrate Photoreceptor Cel...Centrins, a Novel Group of Ca2+-Binding Proteins in Vertebrate Photoreceptor Cells - Madame Curie Bioscience Database
- The Tn916/Tn1545 Family of Conjugative Transposons - Madame Curie Bioscience Dat...The Tn916/Tn1545 Family of Conjugative Transposons - Madame Curie Bioscience Database
- Functions of the Hsp90-Binding FKBP Immunophilins - Madame Curie Bioscience Data...Functions of the Hsp90-Binding FKBP Immunophilins - Madame Curie Bioscience Database
- Important Mammalian Respiratory Allergens Are Lipocalins - Madame Curie Bioscien...Important Mammalian Respiratory Allergens Are Lipocalins - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...