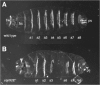NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
Madame Curie Bioscience Database [Internet]. Austin (TX): Landes Bioscience; 2000-2013.
Drosophila melanogaster is a developmentally and genetically highly tractable animal model and many significant advances in the field of signal transduction have been made and elucidated in this system. One example is the JAK/STAT cascade—a pathway conserved throughout evolution and present as a complete canonical pathway in the fly. Studies of pathway mutations, their roles in developmental processes and screens to identify novel pathway regulating loci have identified striking parallels between Drosophila and more complex vertebrate systems. Here we review the major functions of JAK/STAT pathway signalling in Drosophila and examine recent developments likely to lead the field over the coming years.
Introduction
The JAnus Kinase protein and the Signal Transducer and Activator of Transcription represent two core components of the JAK/STAT signal transduction cascade. Originally identified on the basis of the cellular response to interferons,1-3 four JAKs (JAK1,2,3 and TYK2) and seven STATs (STAT1,2,3,4,5a,5b and 6) have since been identified in vertebrate systems along with a large and diverse range of trans-membrane receptors and extracellular ligands (reviewed in ref. 4). In addition to the identification of pathway components, considerable research has also produced insights into the roles that the pathway plays during development, immune responses, hematopoiesis and numerous cancers (reviewed in refs. 5-7).
The JAK/STAT pathway has been conserved throughout evolutionary time and has been studied in many model organisms including mouse, the zebrafish Danio rerio, the fruitfly Drosophila melanogaster, the nematode Caenorhabditis elegans and the slime mould Dictyostelium discoideum. In particular, analysis has focused on mammalian models (often via cell based studies) and the fruit fly Drosophila melanogaster. By contrast to the complexity, inter-dependence and mutual redundancy that often complicates pathway analysis in vertebrates, the situation in Drosophila is significantly more straightforward.
The core components of the canonical JAK/STAT signalling pathway present in Drosophila include three related ligands called Unpaired (Upd), Unpaired 2 (Upd2) and Unpaired 3 (Upd3). Binding of ligand to the predimerised Domeless (Dome) receptor8-10 stimulates signalling and causes the receptor associated JAK tyrosine kinase Hopscotch (Hop)11 to phosphorylate both itself and the cytoplasmic tail of the Dome receptor to create docking sites for the latent STAT92E proteins.12,13 STAT92E is in turn phosphorylated,10 dimerises and translocates into the nucleus where it is capable of binding to a palindromic response element to induce target gene expression.13
The low level of redundancy that results from this relative simplicity, coupled with the availability of a wide range of mutations in pathway components and sophisticated genetic tools allows the pathway to be analysed in detail and has identified a range of developmental processes and immune functions for which pathway signalling is required. Here we introduce the major components and regulators of the Drosophila JAK/STAT pathway and go on to discuss the developmental requirements for the pathway during embryogeneis, larval and adult life. Finally, recent advances, particularly in terms of RNAi based screening and a growing body of evidence suggesting interactions with regulators of chromatin structure, are discussed.
Components of the Drosophila JAK/STAT Pathway
Unpaired
Probably the first mutations in a JAK/STAT pathway component to be described were the outstretched group of regulatory mutations identified on the basis of small eye and held out wing phenotypes.14 Amorphic loss-of-function alleles were subsequently identified during a mutagenesis screen to identify loci required for the proper segmentation of the larval cuticle (Fig. 1). These were classed as a novel group of atypical gap genes on the basis of their striking phenotypes and named Unpaired (Upd) on this basis.15 Subsequent genetic analysis suggested that Upd must represent a non-autonomously acting factor16 while subsequent cloning of the locus showed that Upd encodes a secreted, glycosylated protein that leads to the tyrosine phosphorylation of Hop in cultured Drosophila S2 cells17 and represents the major physiological ligand responsible for the majority of JAK/STAT pathway signalling during many stages of Drosophila development.
Following the sequencing of the Drosophila genome,18 sequence homology searches identified two further upd-like genes located adjacent to upd on the X chromosome.19,20 In vivo analysis of Upd2 suggests that it acts as a semi-redundant ligand being expressed in the embryo in an identical pattern to Upd. While null upd2 mutations are completely rescued by endogenous Upd, the phenotypes arising from the complete loss of upd are only partially compensated by upd2.20 Upd and Upd2 are both secreted and have several potential N-glycosylation sites,20,21 however, they display different biophysical properties with Upd closely associating to the extracellular matrix (ECM) while Upd2 appears to be more freely diffusible20—the significance of this with respect to different in vivo activities is however less clear.
In addition, a third upd-like gene, upd3, has also been identified.19 upd3 is expressed by the hemocytes of adult flies in response to immune challenge after septic injury and is required for JAK/STAT-mediated signalling in the fat body to produce anti-microbial peptides.22 The role of upd3 in other tissues has not yet been established.
Domeless
The JAK/STAT receptor Domeless was identified by a number of independent groups using P-element mutagenesis to look for defects in the posterior spiracle development,8 gut morphology23 or modulation of eye size10 and was ultimately named on the basis of the characteristic dome-shaped posterior spiracle in mutants. Molecularly, Dome resembles a Type 1 cytokine receptor of the JAK associated IL-6 superfamily with homology to gp130 and IL-6R. The extracellular region of the receptor contains five fibronectin type III (FNIII) domains with the first two being homologous to the vertebrate cytokine binding modules (CBM).8,19
Elegant in vivo assays using a bipartite -galactosidase complementation assay have also demonstrated the requirement for Dome homo-dimerisation in multiple developmental tissues as a prerequisite of JAK/STAT pathway signalling,9 while expression of dome itself appears to be at least partly dependent on JAK/STAT pathway activity. Analysis of a dome intronic enhancer region shows clear pathway dependent mesodermal expression20 suggesting the possibility of a positively acting feedback loop of Dome activity in these tissues.
Searches of the Drosophila genome identified CG14225 as a distant homologue of dome. CG14225 is also a transmembrane molecule with two extracellular fibronectin domains forming a CBM and with a striking similarity in domain structure to the vertebrate IL-6Ra receptor.19 As yet no role for CG14225 has been suggested, although it is tempting to speculate on the existence of functional CG14225/Dome as well as Dome/Dome homo- and hetero-dimeric receptor complexes in vivo.
Evidence also exists suggesting noncanonical and potentially Dome-independent, activation of STAT92E under certain gain-of-function scenarios. For example, removal of dCsk, which is a negative regulator of the dSrc tyrosine kinase, leads to cellular overgrowth phenotypes which are strongly suppressed by reducing STAT92E activity.24 It is not, however, clear if the overgrowth phenotype in dCsk mutants, which ultimately leads to elevated Src signalling, is a direct activation of STAT92E or an indirect mechanism that activates JAK/STAT and JNK pathways. A further example of interplay/cross talk between pathways is also suggested by gain-of-function mutations in the Drosophila Torso (Tor) receptor tyrosine kinase, a receptor most homologous to the mammalian PDGFR. Although potentially offering an insight into misregulation of JAK/STAT signalling in human malignancies, the significance of this interaction for normal development is less clear as STAT92E activity is required for gain-of-function mutation of TOR to activate ectopic gene expression, whereas wild-type Tor signalling appears to be mediated by the Ras/Raf/MAPK signalling pathway.25,26
Hopscotch
Initially identified on the basis of a segmentation phenotype (Fig. 1) strikingly similar to that of Upd, mutations in hopscotch (hop) also reduce the size of imaginal discs, ovaries, testis, hindgut, the foregut imaginal ring and proliferation regions of the brain.27 Following its molecular characterisation, hop was found to encode a nonreceptor tyrosine kinase of the JAK family11 and is most similar to mammalian JAK1 and 2.10 Hop contains one functional and one pseudo-kinase domain, a SH2-like domain and a FERM domain.19 Intriguingly, subsequent genetic study identified the long standing l(1)Tumours lethal (Tum-l) mutation as a temperature sensitive dominant gain-of-function allele of hop28 (henceforth termed hopTum-l). Intriguingly, these mutations result in the over-proliferation of Drosophila blood cells and the formation of melanotic blood cell tumours. Molecular characterisation of hopTum-l and a second gain-of-function allele hopT42, identified lesions in the JH2 domain which result in constitutive kinase activity29,30—findings with particular parallels to the more recent identification of JAK2 activating V617F mutations recovered in a wide range of human meyoproliferative diseases.31-33
STAT92E
The transcription factor of the Drosophila JAK/STAT pathway, STAT92E, was identified by two independent screens, a cDNA library screening approach on the basis of homology to human STATs13 and a genetic approach screening for P-elements producing segmentation defects similar to upd and hop mutants (Fig. 1).12 STAT92E is most similar to mammalian STAT3 and STAT5 and has an essentially conserved domain structure including an N-terminal domain, coiled-coil region, a DNA binding domain, SH2 domain and conserved tyrosine residue likely to represent the target of Hop phosphorylation. Following activation Drosophila STAT92E has been shown to translocate to the nucleus10,22,34-36 and bind to a palindromic DNA target sequence13 to activate transcription.34,37 In addition, stat92E is expressed as a number of splice variants including an N-terminal 133 amino acid truncated form termed ΔNSTAT92E suggested to function as a dominant negative regulator of JAK/STAT pathway signalling in vivo.38
Intriguingly, recent developments also suggest other roles for STAT92E in addition to its canonical role as a pathway activated transcription factor. For example, mutational analysis of STAT92E have separated DNA binding from the transcriptional activation activities34 while recent studies have also shown a role for unphosphorylated, DNA-associated STAT92E in regulation of heterochromatin protein 1 (HP1) and the progression of heterochromatinisation39 (see also below).
Negative Pathway Regulators
SOCS36E
The socs genes represent the best-characterised negative regulators of the JAK/STAT pathway, both in mammals and Drosophila. Many are themselves target genes of pathway signalling and form a potent negative feedback loop which functions to down-regulate JAK/STAT pathway activity. Three socs-like genes have been identified in the Drosophila genome, socs16D, socs36E and socs44A, all of which contain an SH2 domain and SOCS box (SB). SOCS36E is most similar to mammalian SOCS5 with 29.7% identity whereas SOCS44A and SOCS16D are most similar to mammalian SOCS6 and 7. Of the three Drosophila homologues, SOCS36E is the only JAK/STAT pathway repressor that is also a direct target gene of the pathway.35,37,40,41 In addition SOCS36E is not only a negative regulator of JAK/STAT pathway activity,35,37 but has also been reported to suppress EGFR signalling activity in the wing disc suggesting that it can interact with multiple pathways.41 Although SOCS44A is not a direct target of the JAK/STAT pathway, it can inhibit its activity in some tissues while it also appears to up-regulate EGFR signalling.42 It is not yet clear, however, if this is a direct or indirect effect. The function of SOCS16D has yet to be characterised.
dPIAS/Su(var)2-10
PIAS proteins were originally identified in vertebrate systems and termed Protein Inhibitors of Activated STAT. PIAS1 and 3 physically interact with STAT1 and 3 respectively and their over-expression in transfected cells inhibits STAT-induced transcription.43,44 However, the physiological relevance of PIAS in vivo was first established with its Drosophila homolog dPIAS (originally known as zimp45). Reduction or over-expression of dPIAS in flies leads to increases and decreases in JAK/STAT activity, respectively, suggesting that dPIAS acts as a negative regulator of the pathway in vivo.37,46 Intriguingly, more recent studies in mammalian systems suggest that PIAS proteins have diverse functions including roles as SUMO-ligases, while genetic studies in Drosophila have shown that dPIAS is in fact allelic to the Su(var)2-10 locus. Su(var)2-10 is a modifier of the position effect mottling of eye pigmentation in the adult fly eye thought to represent a readout of heterochromatinisation with mutants in Su(var)2-10 having defects in nuclear structure and chromatin segregation.47 It is not yet however clear if these STAT regulation and Su(var) effects are caused by a failure of PIAS-mediated SUMOlation .
Ken and Barbie
The interaction of mutations in ken and barbie (ken) with the JAK/STAT pathway was first shown in a genetic interaction screen suggesting that ken acts as a negative regulator of the pathway.48 The ken gene encodes a DNA-binding protein containing three zinc fingers and an N-terminal BTB/POZ domain commonly found in transcriptional repressors and appears to be a homologue of human B-cell lymphoma 6 (BCL6).49,50 Strikingly, the core DNA binding sequence of Ken identified in vitro overlaps half of the palindromic STAT92E DNA binding sequence.49 Luciferase reporter assays carried out in cultured cells and using sites to which either STAT92E and Ken or only STAT92E can bind show that Ken acts as a repressor of the JAK/STAT pathway. Ken, however, only suppresses a subset of JAK/STAT target genes in vivo with the JAK/STAT targets vvl, trh and kni being repressed while socs36E expression is not affected.49 This result underlines the observation that modulation of signalling pathways via subtle changes to transcription factor binding sites can have significant effects on cell-specific transcriptional responses to ligand stimulation. Intriguingly, in mammalian systems BCL6 has also been shown to bind to STAT6 DNA binding sites where it can act as a repressor of STAT6 dependent target gene expression51,52—however a potential link between the modulation of JAK/STAT signalling and the association of BCL6 with B-cell lymphomas remains to be investigated.
PTP61F
ptp61F is one of the 28 predicted protein tyrosine phosphatases encoded in the fly genome53 and was identified as a JAK/STAT pathway regulator in two independent genome-wide RNAi screens.35,37 PTP61F is a homologue of the human phospho-tyrosine phosphatase B1 (PTPB1) and is implicated in the de-phosphorylation of both JAKs and STATs in both the cytoplasm and nucleus of mammalian cells.54 However some confusion remains regarding the true in vivo substrate of PTP61F that regulates JAK/STAT signalling. A mechanism invoking direct de-phosphorylation of activated, nuclear localised STAT92E has been proposed by one group37 while knock down by RNAi results in increased phosphorylation of both Drosophila Hop and STAT92E.35 Even more confusingly, recent proteomics approaches have failed to identify interaction of any JAK/STAT pathway components to a substrate trapping variant of PTP61F.55 As such, the precise mechanism of action remains unclear. None the less, in vivo studies underline the repressing activity of PTP61F on the JAK/STAT pathway35,37 and its embryonic expression in a pattern highly reminiscent of upd indicates that ptp61F expression is likely to be a target of the JAK/STAT pathway suggesting the existence of yet another negative feedback loop level of pathway regulation.35
Developmental Roles
Embryonic Development
The Drosophila JAK/STAT pathway plays a number of key roles at multiple stages during embryogenesis being activated in diverse tissues and at multiple stages. Its roles start with sex determination and segmentation and subsequently the formation of the tracheal pits, elongation of intestinal tracks and formation of the posterior spiracles. Other, as yet less well defined roles include a role in the normal development of axons within the central nervous system56 and embryonic dorsal/ventral patterning.57 Here we will discuss the better-studied roles of the JAK/STAT pathway in each tissue in greater detail.
Sex Determination
In Drosophila, sex is determined by the ratio of X chromosome(s) to autosomes. This ratio information is supported by X-linked signal elements (XSE), which turn on the expression of sex-lethal (sxl) in females but not in males. Upd, among others, is one of these XSEs.58,59 While other XSEs induce transcription of sxl by directly binding to its promoter, Upd is a secreted ligand, which must activate Sxl promoter indirectly. In support of this, the sxl promoter contains two STAT92E binding sites and loss-of-function of hop and stat92E mutations reduce sxl expression. Intriguingly and consistent with this model, genome wide RNAi screening data also indicates that the Drosophila X-chromosome contains a number of positively activating JAK/STAT pathway components and regulators but no identified negatively acting factors.37
Segmentation
Study of the processes regulating the segmentation of the Drosophila embryo has a long history15 and elucidation of this process has provided profound insights into multiple developmental processes. A combination of molecular, genetic and embryological approaches has led to a hierarchical model describing how a segmented pattern of three thoracic and eight abdominal segments are generated along the anterior-posterior axis. Mutations removing upd, dome, hop and stat92E all lead to deletion or fusion of the fourth and fifth abdominal segment and partial loss of the eigth abdominal segment, as well as defects in the head skeleton and posterior spiracles (Fig. 1).11,12,27 This distinctive phenotype, however, is not consistent with those of the classical maternal, gap, pair rule or segment polarity phenotypic classes (reviewed in ref. 60). Rather, mutants of the JAK/STAT pathway show an atypical gap gene phenotype due to a localised reduction in expression of the pair-rule genes even-skipped (eve), runt and fushi tarazu11 Direct molecular evidence to support this finding has been established for eve where two crucial STAT92E binding sites are located within the eve stripe 3-7 promoter.11,61
Trachea
The tracheal system supplies oxygen to all organs of the fly via a network of air filled tubules radiating throughout the body with the posterior spiracles representing the external opening during larval stages. The embryonic tracheal system is an epithelial tubular network established from ectodermal precursor cells. During embryonic development tracheal pits are formed which define a population of cells that divide, migrate and fuse to form the complete tubular network. The JAK/STAT ligand upd is expressed in tracheal pits17 and activates its downstream transcription factor STAT92E.56 Mutants that lack maternal and zygotic contribution of hop or stat92E do not form any tracheal system.8,27 Tracheal pits and posterior spiracles are marked at early stages of embryogenesis by trachealess (trh), knirps and ventral veins lacking (vvl), expression of which is lost in JAK/STAT pathway mutants.8 Trh is required early for trachea specification and later to promote invagination to form tubes. The JAK/STAT pathway therefore seems to play a dual role in the trachea, namely activating downstream target genes, which are crucial for cell movement and elongation as well as specifying these specialised cells for invagination.62
Gut
The Drosophila hindgut is a single-layered ectodermally derived epithelium which elongates and narrows during a 10 h period of late embryogenesis (reviewed in ref. 63) and provides an excellent system to study cell-shape changes and cell rearrangements. The process of hindgut elongation during embryogenesis requires polarized cell rearrangements, which are supported by local expression of signalling molecules including hedgehog, wingless and upd.64-66 Upstream of these pathways, transcription factors including drumstick (drm), bowel (bowl) and lines (lin) are required. Whereas in drm and bowl mutants expression of upd is reduced, in lin mutants upd expression is expanded suggesting that localised Upd protein might be an important cue for hindgut cells.67 Inactivation of the JAK/STAT pathway leads to shorter but wider hindgut phenotype, a change not due to reduced cell number, but rather due to the loss of spatially localised JAK/STAT signalling required to trigger cellular rearrangement within the hindgut via a process of convergent extension.67
In Drosophila, the activity of Hedgehog, Wingless and the TGF-β homologue Decapentaplegic (Dpp) are also required for the development of the foregut-associated proventriculus.68,69 This organ regulates the passage of food into the midgut and is composed of endodermal, mesodermal and ectodermal layers. Localised JAK/STAT and Notch signalling are required for proper cellular migratory events in the foregut, which also involve cytoskeletal reorganisation.23,70
Ultimately, understanding of how JAK/STAT pathway signalling is able to trigger and control cellular movement and rearrangement, not only in the hind gut and the foregut, but also as part of tracheal development and border cell migration (discussed below), will be an important avenue for future research.
Larval Development
Imaginal Discs
The JAK/STAT signalling pathway plays several important roles in the development of both the eye and wing imaginal discs. Imaginal cells are set aside during embryogenesis and proliferate throughout larval life before differentiating and everting during pupal stages to form a large proportion of the adult fly. One of the first roles for JAK/STAT signalling in this process is a requirement for the pathway in cellular proliferation. Regulatory loss-of-function alleles of Upd with small eye phenotypes are amongst the first pathway mutations identified14 and constitutive expression of dominant negative pathway components in the eye primordia are sufficient to significantly reduce eye size.48 Conversely, ectopic pathway activation within the developing eye imaginal disc leads to cellular over-proliferation ahead of the morphogenetic furrow—a zone of multipotent eye progenitor cells.48,71 This JAK/STAT pathway-induced over proliferation and the large increase in adult eye size that results, is dependent on the normal diploid dose of downstream pathway components. As a consequence, mutation of a single copy of positively acting pathway components is sufficient to cause a relative decrease in eye over-proliferation—an effect subsequently used to undertake a number of genetic screens for previously unidentified pathway regulators.48,72
A detailed in vivo analysis of the role of JAK/STAT pathway signalling in the regulation of cellular proliferation has also been undertaken in the developing wing imaginal disc.73 Intriguingly, clonally related groups of marked cells mutant for stat92E loss-of-function alleles show different proliferative profiles at differing developmental stages. During early larval stages, loss of the pathway reduces the rate of cellular proliferation. This is consistent with our understanding in the eye and genetic studies implicating the Drosophila Cdk4 and Cyclin D-cell cycle proteins as physical interaction partners of STAT92E.74 However, during later stages, when upd expression in the wing disc is more localised, removal of stat92E causes an increase in the rate of cellular proliferation.73 While the mechanisms underlying this change in pathway function remain to be determined, it is intriguing that the single Drosophila stat92E locus can mediate both pro- and anti-proliferative functions in the same tissue during the course of development.
In addition to roles in regulating cellular proliferation, JAK/STAT pathway signalling is also an important regulator of a number of additional stages of eye imaginal disc development. It has recently been shown that the activation of STAT92E in the eye is required to repress wingless (wg) expression and so allows the proper initiation of the morphogenetic furrow,75,76 while at later stages the JAK/STAT pathway is also required for correct ommatidial rotation77 via a process mediated (at least in part) by the pathway dependent expression of the planar polarity regulator four-jointed.78 In addition, repression of both wg and the Dpp expression in the developing leg and antennal discs has also been shown to be at least partly under the control of JAK/STAT signalling where the pathway regulates the proximal distal growth of the appendages.79
Intriguingly, expression of upd appears to be under the control of the Notch signalling pathway in multiple developmental contexts, including in the eye,80,81 the embryonic foregut23 and ovarian follicle cells.82 Indeed, ectopic activation of Notch signalling in the eye caused by mutations in endocytic pathway components such as vps2583,84 and tsg10185 lead to Upd mediated non-autonomous eye overgrowth. As such, it is clear that Upd/JAK/STAT signalling processes are targets of upstream factors and regulators of diverse downstream signalling pathways at many stages of development. The recent development of sensitive in vivo reporters of JAK/STAT pathway activity86 will no doubt help to unravel these complex interactions in the future.
Hematopoiesis
Drosophila blood cells, termed hemocytes, perform two significant functions. They search for and engulf pathogens and apoptotic cell debris (especially during pupal stages) and they monitor the environment for bacterial, fungal, parasitic and viral infection in order to signal to the fat body to mount appropriate innate immune responses (reviewed in refs. 87, 88).
The three types of blood cells constitute macrophage like plasmatocytes (90-95% of hemocytes), crystal cells (~5%) and lamellocytes. Lamellocytes are large cells that are rarely observed in healthy larva, however large numbers are induced in larval stages upon challenge with parasitic wasp eggs89 where they function to encapsulate objects that are too large to be engulfed by plasmatocytes.90
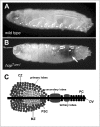
Several signal transduction pathways, including the JAK/STAT, Toll, Ras/Raf, Hedgehog and Notch cascades, have been shown to be crucial for Drosophila hematopoiesis.29,91-95 In particular, two temperature sensitive gain-of-function mutations of the Drosophila Hop kinase, named hopTum-l 28,29 and hopT42,30 illustrate the key role played by JAK/STAT signalling in haemocyte development. At low temperatures, where hopTum-l is only moderately over active, larvae contain 5-20 fold more plasmatocytes than wild type,93 however, when raised at higher temperatures the more strongly signalling gain-of-function HopTum-l mutation drives haemocyte differentiation into lamellocytes, which contribute to auto-encapsulation and black melanotic tumours (Fig. 2A and B).93 The formation of HopTum-l-induced tumours can be suppressed by the removal of one copy of downstream pathway components such as stat92E or over expression of dPIAS,37 while transplantation of the larval haematopoietic organ from a HopTum-l mutant into a wild-type host results in the appearance of melanotic masses, implicating invasive properties of the tumours.96
The primary source of larval hemocytes is the haematopoietic organ or lymph gland, a lobed structure located along the dorsal vessel (Fig. 2C). The most anterior lobe of the lymph gland is divided into three distinct regions, the posterior signalling centre (PSC), the medullary zone (MZ), which is marked by the expression of the Domeless receptor and the outer cortical zone. The PCS acts as a signalling centre by expressing the ligands of a number of signalling pathways including Hedgehog,94 the Notch ligand Serrate (Ser)97 and probably Upd398 which is thought to induce the JAK/STAT pathway in the MZ to maintain the undifferentiated state of pro-hemocytes within this region.99
Although the precise relationship between Hedgehog, Notch and JAK/STAT pathway signalling in controlling pro-haemocyte development and maintenance is not yet clear, thin cytoplasmic fillopodial extensions from the PSC suggest that direct cellular contact may be involved.94,99
While the JAK/STAT pathway is important for lamellocyte differentiation, haemocyte over proliferation has been described to be STAT92E independent.29 This suggests that hop may crosstalk with other pathways to mediate plasmatocyte over-proliferation observed in hopTum-l mutants at low temperatures.93 Indeed, Hop has been shown to physically interact and activate both STAT92E and dRaf. dRaf is a key component of the Ras/Raf/Erk pathway, a cascade that has itself been shown to play a key role in haemocyte proliferation93,100 with the added twist that expression of the draf gene is itself at least partly under the control of the STAT92E transcription factor in vivo.101 Overall, it seems likely that both Ras/Raf and JAK/STAT signalling pathways play key roles in Drosophila blood cell development although the details of a model in which the differentiation of hemocytes into lamellocytes via the JAK/STAT pathway and cell proliferation via Erk mediated signalling remain to be determined.
Adult Stages
Immunity
Although the innate insect immune response to bacterial and fungal infections involves the signalling pathways Toll and Imd (reviewed in ref. 102), evidence that the JAK/STAT pathway also plays a role in insect immunity was shown in the mosquito Anopheles ganbiae where AgSTAT translocates into the nucleus of fat body cells in response to bacterial infection.103 In Drosophila, STAT92E also translocates into the nucleus of fat body cells upon immune challenge, the result being expression of several anti-microbial peptides including the Tep and Tot protein families.22
Tep1 is one of the four members of the Tep family each of which contain thiolester motifs and have high similarity to thiolester-containing proteins of the complement C3/alpha2-macroglobulin super family. Tep1 expression is highly induced in fat body cells upon immune challenge.104 This expression is dramatically reduced in hypomorphic hop loss-of-function mutant larvae and is constitutive in HopTum-l individuals. In vertebrates, the complement system mediates inflammatory reactions. Activation of the C system can occur by the lectin pathway, among others, leading to the development of the complement C3 protein.105 Intriguingly, the JAK/STAT ligand upd has been suggested to be weakly homologous to leptin106 although the potential significance of this is as yet unclear.
The Tot family encodes eight small peptides, whose transcription is activated in response to bacterial challenge or environmental stress, such as UV-radiation, high temperature, or oxidizing agents.22,107,108 TotA expression is reduced in challenged flies carrying hypomorphic hop loss-of-function alleles and is dependent on the Domeless receptor in the fat body. Flies carrying an upd hypomorphic allele or upd2 null allele continue to show a normal totA expression in response to challenge. However upd3 expression in the hemocytes (but not in the fat body) is necessary to activate the JAK/STAT pathway in the fat body leading to totA expression.22 The closely related TotC and TotM are also controlled by JAK/STAT pathway during septic injury.22,109
An innate defence against viral infection also seems to involve the Drosophila JAK/STAT pathway, which is required, but not sufficient, for the antiviral response. Infection of flies with Drosophila C virus induces the transcription of virus-induced RNA 1 (vir-1). Its expression requires JAK/STAT pathway activation and its promoter contains STAT92E DNA binding sites to which the transcription factor is able to bind, while expression is diminished in hypomorphic hop loss-of-function backgrounds.110
Ultimately, although our understanding of the interplay between the Toll, Imd and JAK/STAT pathways in triggering the appropriate responses to sterile and septic injuries, as well as parasitic and viral infections is already partly understood,109 this area will no doubt represent an important field for future research.
Stem Cell Niches
The importance of the stem cells niche, a microenvironment that provides external signals to maintain surrounding stem cells, is becoming increasingly appreciated across many species (reviewed in refs. 111-114). Intensive work on niches reveal that while primary signals are crucial for germline stem cell (GSC) homeostasis, hierarchies among multiple pathways are also important (reviewed in ref. 115). Adult stem cell niche studies in invertebrates have shed light on the nature of the signals which mediate stem cell regeneration capacity in vivo with the Drosophila GSCs representing one of the best studied adult stem cell niches. In males, tightly packed somatic hub cells at the tip of the testis signal to adjacent GSCs by producing the Upd ligand.116-118 Asymmetric division of stem cells produces one stem cell close to the hub and one displaced cell further away; secretion of Upd from the hub activates the JAK/STAT pathway, promoting self-renewal in the nearby GSC, while the one further from the source of ligand subsequently differentiates,119,120 a process that is further supported by the MAP kinase pathway.121,122 Consistent with this model, constitutive JAK/STAT pathway activation results in a testis filled with stem cell-like cells, while loss of pathway components causes the differentiation and loss of the GSCs.117,118,123 Reductions in the levels of upd and cadherin expression in the niche cells over time correlates with a decrease in the number of GSCs, indicating that aging of the stem cell niche may well be a factor in decreased stem cell activity.124 In addition to Upd, Dpp signalling is also required for male GSC niche.116,125
The female ovary consists of multiple ovarioles each of which consists of strings of individual egg chambers at progressively more mature developmental stages. The youngest, most anterior end of the ovariole contains two populations of stem cells, the GSC and the somatic escort stem cells (ESC). It was long thought that the maintenance of GSC is performed through a microenvironment formed by somatic cap cells which secrete Dpp to maintain the niche.126 However, several studies have now shown that the JAK/STAT pathway lies at the top of a hierarchical signalling model. The pathway not only positively regulates Dpp expression in the cap cells127,128 but is also needed in ESCs to maintain the structural integrity of the germarium.129 The requirement of JAK/STAT activity in the GSC niche of both sexes is however different. Whereas cell autonomous JAK/STAT activity is required in male GSCs, JAK/STAT signalling in female ovary is needed only for the maintenance of Dpp signalling in the cap cells and is dispensable for female GSC self renewal.
In addition to the relatively well characterised roles in germ line stem cell maintenance, the activity of JAK/STAT pathway signalling is also required for the maintenance of haematopoietic stem cells in the lymph gland99 and multipotent stem cells in the malpighian tubules (Drosophila kidney).130 These findings suggest a recurring theme in which pathway activity is a central component in the establishment and maintenance of stem cell niche environments in vivo. A potential link between this role and the recently popular concept of ‘cancer stem cells’131,132 is particularly intriguing, especially given the prevalence of inappropriate JAK/STAT pathway activation in a range of human malignancies.133
Oogenesis
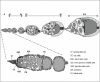
As discussed above, JAK/STAT signalling is required to maintain the stem cells that will ultimately generate the somatic support cells required for normal oogenesis.129 The Drosophila ovary consists of multiple ovarioles, each of which contains a string of developing egg chambers. Egg chambers are surrounded by a monolayer of somatic epithelial follicle cells, which undergo a highly stereotypical series of movements to enclose the developing oocyte (Fig. 3). Epithelial cells in the Drosophila ovary offer a powerful system to model the study of the development of invasive behaviour and have been shown to actively require JAK/STAT signalling for this process.134 These epithelial follicle cells differentiate into three distinct cell populations135,136 to establish the egg chamber: polar cells at each end of the egg chamber, stalk cells connecting individual chambers and main-body follicle cells overlying the egg chamber itself. Induction of Notch signalling from the germline cells to neighbouring follicle cells induces polar cell fate.137-139 However, after stalk induction, Upd signalling extends further resulting in a gradient of JAK/STAT signalling across the main-body follicle cells located at the posterior end of the ooctye. Upd ligand secreted from the anterior polar cells signals to adjacent follicle cells defining their fate to border cells, which delaminate from their normal position, undergo an epithelial to mesenchymal transition (EMT) and acquire the ability to migrate between the supporting nurse cells to contact the maturing oocyte (Fig. 3 - dark cells at stage 9-10).134,140 Over-expressing Upd or Hop is sufficient to recruit surrounding follicle cells into a border cell fate, which then exhibit invasive ‘metastatic’ behaviour.134,140
This final requirement of the JAK/STAT pathway for the EMT and movement of polar cells has parallels in other Drosophila tissues such as the tracheal pits and gut where JAK/STAT dependent cell movements are crucial to normal development23 (reviewed in ref. 141). The significance of the recurring theme of JAK/STAT pathway mediated cellular movement may be of particular significance given the poor prognosis of patients with metastatic human cancers. Investigation of the role for JAK/STAT signalling in this process represents a potentially important theme for future research.
Future Directions
Chemical and Genome-Wide RNAi Screens
One of the greatest strengths of the Drosophila experimental system is its tractability as a ‘screenable’ genetic tool. Such screening approaches have also been applied to the study of JAK/STAT pathway signalling in flies and a number of classical genetic screens have been undertaken.48,72 These have identified a number of novel pathway regulators including the transcriptional repressor ken and Barbie.49 The publication of the Drosophila genome in 200018 and the discovery of RNA interference (RNAi) as a tool for genome-wide screening142,143 have further widened the possibilities for screening. Genome wide libraries of double stranded (ds)RNAs targeting all predicted Drosophila transcripts have been generated144 and large scale cell-based screening has made possible forward genetic studies to identify genes that modulate signalling pathways (reviewed in refs. 145, 146).
Two genome-wide, cell-based screens for JAK/STAT pathway modulating loci have been undertaken in recent years.35,37 Using STAT92E-responsive luciferase reporters in cultured Drosophila cells, a range of both positively acting genes, whose knock down results in a decrease in JAK/STAT pathway reporter activity and negatively acting loci, whose knock down results in an increase in signalling, have been identified. Following cell-based validation, both screens identified around 100 enhancer or suppressor loci whose knock down was sufficient to modulate JAK/STAT pathway activity. These included core pathway components, the pathway phosphatase ptp61F (described above) and a number of genes whose human homologues have been previously associated with leukaemia,35,37 (reviewed in ref. 147). While the further analysis of these JAK/STAT pathway modulating loci, both in Drosophila and in vertebrate model systems, will undoubtedly represent an important future research direction, it is also likely that further genome-wide RNAi screens will be undertaken as the screening technology itself matures. Ultimately, the increasing ease with which such screens can be performed is likely to allow ‘smaller’ questions to be addressed. For example, high-content visual screens for loci that change the sub cellular localisation of STAT92E, or screens to identify the factors required for receptor predimerisation9 can be devised. Screens to identify factors required in Upd-secreting cells versus receiving cells or to identify differences in the factors that interact with the three Upd-like ligands are all possible. As such, assays will not only identify pathway interacting genes, but will start to characterise them on the basis of their function as well.148 Such functional screening, in combination with the in vivo developmental genetic analysis of loci with respect to the known roles of JAK/STAT pathway signalling, are likely to represent a major future endeavour.
Another potential approach to identify regulators of JAK/STAT signalling is the use of chemical genetic studies utilising small molecules that modulate protein function in cell-based assays.149 Although this has yet to be undertaken for Drosophila JAK/STAT signalling, complementary approaches combining chemical and genome-wide RNAi screens have the potential to rapidly identify the target through which a small molecule inhibitor of pathway signalling acts. By carrying out essentially identical small molecule and RNAi screens, the loss of protein activity elicited chemically should have similar readouts to RNAi depletion of the mRNA encoding the same protein. Given that the target of the dsRNA is known, the likely mechanism of action of small molecule inhibitors in whole cell conditions can be rapidly ascertained. Ultimately, practical experience will be needed to determine exactly how effective such cell scale drug discovery efforts prove to be (reviewed in ref. 150).
JAK/STAT Pathway and Chromatin Structure
Under normal conditions, the DNA of each cell is ‘imprinted’ with an epigenetic code thought to ‘shut down’ large proportions of the genome not relevant to the current cell fate (reviewed in refs. 151, 152). In particular, the formation and maintenance of a balance between ‘open’ euchromatin and essentially silenced heterochromatin is likely to be important in this process and its disruption could well be a fundamental factor in the development of cancers.153
A number of threads of evidence seem to suggest that the JAK/STAT pathway may well play a role in regulating such epigenetic heterochromatinisation effects. Firstly, the Drosophila PIAS homologue (discussed above) has been shown to be allelic to the Su(var)2-10 locus—a gene identified on the basis of its ability to prevent the heterochromatic silencing of eye pigmentation during development.47 Secondly, a number of putative chromatin modifying genes were identified in the genome wide RNAi screens37 and the human homologues of some of these, such as the Histone deacetylases MYST3/MOZ, have previously been implicated in human leukaemias.154 Thirdly, the nucleosome remodelling factor NURF301, has been shown to be a negative regulator of the tumour phenotype induced by HopTum-l.155,156 Transcript profiling of NURF and HopTum-l mutants identifies overlapping sets of genes whose expression is likely to be regulated by an overlapping STAT92E and Ken binding sites in their promoters.157 Furthermore, Ken physically binds to NURF recruiting it to STAT binding sites and leading to repression of JAK/STAT target genes.
Finally, a genetic interaction screen carried out to identify loci that enhance or reduce the melanotic tumour phenotype induced by HopTum-l has also been undertaken. Intriguingly, a major class of mutated genes identified in this screen also represent putative chromatin modification proteins.156 Two of these mutated genes include Su(var)3-9 which catalyzes the methylation of Histone3 at Lys9 and Heterochromatin Protein 1 (HP1) which initiates heterochromatin formation and spreading. Whilst removal of a single copy of these loci leads to an increase in HopTum-l-mediated tumour formation, increasing their expression levels suppresses tumourogenesis suggesting that heterochromatin formation and JAK/STAT pathway signalling may be linked.
More recently, a follow up analysis of HP1 in this context has demonstrated an initially unexpected interaction of HP1 with apparently ‘inactive’ STAT92E.39 In this potentially highly significant report, Shi and colleagues report that STAT92E is directly required to control HP1 distribution, heterochromatin stability and Histone3 Lysine9 methylation. Intriguingly, this effect is mediated by unphosphorylated ‘transcriptionally inactive’ STAT92E. Reductions in the gene dosage of STAT92E or its recruitment to canonical signalling by phosphorylation disrupts this complex, causes HP1 displacement and subsequently leads to the destabilisation of heterochromatin.39 As such, constitutive activation of STATs could potentially lead not only to the up-regulation of pathway target genes via the ‘traditional’ signalling cascade, but would simultaneously destabilise heterochromatin and so indirectly up-regulate the genes located in these normally silenced regions.
Taken together, these multiple indications of synergy between chromatin remodelling factors such as NURF, HP1 and Su(var) genes and the STAT transcription factor itself represents a significant development in our understanding of how JAK/STAT pathway signalling exerts its diverse effects in vivo. This emerging field will undoubtedly represent a very important area for future research both in Drosophila and vertebrate models.
Acknowledgements
The authors wish to thank Mary Ann Price and Steve Brown for valuable comments on the manuscript. SB is supported by the Deutsche Forschungs Gemeinschaft (DFG) while MPZ is a Cancer Research-UK Senior Cancer Research Fellow and a member of the MRC Centre for Developmental and Biomedical Genetics.
References
- 1.
- Lutticken C, Wegenka UM, Yuan J. et al. Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130. Science. 1994;263(5143):89–92. [PubMed: 8272872]
- 2.
- Stahl N, Boulton TG, Farruggella T. et al. Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL-6 beta receptor components. Science. 1994;263(5143):92–5. [PubMed: 8272873]
- 3.
- Darnell JE Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264(5164):1415–21. [PubMed: 8197455]
- 4.
- Leonard WJ. Role of jak kinases and STATs in cytokine signal transduction. Int J Hematol. 2001;73(3):271–277. [PubMed: 11345192]
- 5.
- Benekli M, Baer MR, Baumann H. et al. Signal transducer and activator of transcription proteins in leukemias. Blood. 2003;101(8):2940–54. [PubMed: 12480704]
- 6.
- Bowman T, Garcia R, Turkson J. et al. STATs in oncogenesis. Oncogene. 2000;19(21):2474–2488. [PubMed: 10851046]
- 7.
- Rane SG, Reddy EP. JAKs, STATs and Src kinases in hematopoiesis. Oncogene. 2002;21(21):3334–58. [PubMed: 12032773]
- 8.
- Brown S, Hu N, Hombria JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. 2001;11(21):1700–5. [PubMed: 11696329]
- 9.
- Brown S, Hu N, Hombria JC. Novel level of signalling control in the JAK/STAT pathway revealed by in situ visualisation of protein-protein interaction during Drosophila development. Development. 2003;130(14):3077–84. [PubMed: 12783781]
- 10.
- Chen HW, Chen X, Oh SW. et al. Mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 2002;16(3):388–98. [PMC free article: PMC155335] [PubMed: 11825879]
- 11.
- Binari RN. Perrimon Stripe-specific regulation of pair-rule genes by hopscotch, a putative Jak family tyrosine kinase in Drosophila. Genes Dev. 1994;8(3):300–12. [PubMed: 8314084]
- 12.
- Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84(3):411–9. [PubMed: 8608595]
- 13.
- Yan R, Small S, Desplan C. et al. Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84(3):421–30. [PubMed: 8608596]
- 14.
- Mller HJ. Types of visible variations induced by X-rays in Drosophila. J Genet. 1930;22(3):299–334.
- 15.
- Wieschaus E, Nusslein-Volhard C, Jurgens G. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster III. Zygotic loci on the X-chromosome and fourth chromosome. Roux Arch dev Biol. 1984;193:296–307. [PubMed: 28305339]
- 16.
- Gergen JP, Wieschaus EF. Localized requirements for gene activity in segmentation of Drosophila embryos: analysis of armadillo, fused, giant and unpaired mutations in mosaic embryos. Roux Arch dev Biol. 1986;195(1):49–62. [PubMed: 28305277]
- 17.
- Harrison DA, McCoon PE, Binari R. et al. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12(20):3252–63. [PMC free article: PMC317220] [PubMed: 9784499]
- 18.
- Adams MD, Celniker SE, Holt RA. et al. The genome sequence of Drosophila melanogaster. Science. 2000;287(5461):2185–95. [PubMed: 10731132]
- 19.
- Hombria JC, Brown S. The fertile field of Drosophila Jak/STAT signalling. Curr Biol. 2002;12(16):R569–75. [PubMed: 12194841]
- 20.
- Hombria JC, Brown S, Hader S. et al. Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Dev Biol. 2005;288(2):420–33. [PubMed: 16277982]
- 21.
- Gilbert MM, Weaver BK, Gergen JP. et al. A novel functional activator of the Drosophila JAK/STAT pathway, unpaired2, is revealed by an in vivo reporter of pathway activation. Mech Dev. 2005;122(7-8):939–48. [PubMed: 15925495]
- 22.
- Agaisse H, Petersen UM, Boutros M. et al. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev Cell. 2003;5(3):441–50. [PubMed: 12967563]
- 23.
- Josten F, Fuss B, Feix M. et al. Cooperation of JAK/STAT and Notch signaling in the Drosophila foregut. Dev Biol. 2004;257(1):181–189. [PubMed: 14975725]
- 24.
- Read RD, Bach EA, Cagan RL. Drosophila C-terminal Src kinase negatively regulates organ growth and cell proliferation through inhibition of the Src, Jun N-terminal kinase and STAT pathways. Mol Cell Biol. 2004;24(15):6676–89. [PMC free article: PMC444864] [PubMed: 15254235]
- 25.
- Li WX, Agaisse H, Mathey-Prevot B. et al. Differential requirement for STAT by gain-of-function and wild-type receptor tyrosine kinase Torso in Drosophila. Development. 2002;129(18):4241–8. [PMC free article: PMC3090254] [PubMed: 12183376]
- 26.
- Li J, Li WX. Drosophila gain-of-function mutant RTK torso triggers ectopic Dpp and STAT signaling. Genetics. 2003;164(1):247–58. [PMC free article: PMC1462547] [PubMed: 12750336]
- 27.
- Perrimon N, Mahowald AP. l(1)hopscotch, A larval-pupal zygotic lethal with a specific maternal effect on segmentation in Drosophila. Dev Biol. 1986;118(1):28–41. [PubMed: 3095163]
- 28.
- Hanratty WP, Dearolf CR. The Drosophila Tumorous-lethal hematopoietic oncogene is a dominant mutation in the hopscotch locus. Mol Gen Genet. 1993;238(1-2):33–7. [PubMed: 8479437]
- 29.
- Luo H, Hanratty WP, Dearolf CR. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. Embo J. 1995;14(7):1412–20. [PMC free article: PMC398227] [PubMed: 7729418]
- 30.
- Luo H, Rose P, Barber D. et al. Mutation in the Jak kinase JH2 domain hyperactivates Drosophila and mammalian Jak-Stat pathways. Mol Cell Biol. 1997;17(3):1562–71. [PMC free article: PMC231882] [PubMed: 9032284]
- 31.
- Jones AV, Kreil S, Zoi K. et al. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood. 2005;106(6):2162–8. [PubMed: 15920007]
- 32.
- Kralovics R, Passamonti F, Buser AS. et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352(17):1779–90. [PubMed: 15858187]
- 33.
- James C, Ugo V, Le Couedic JP. et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434(7037):1144–8. [PubMed: 15793561]
- 34.
- Karsten P, Plischke I, Perrimon N. et al. Mutational analysis reveals separable DNA binding and trans-activation of Drosophila STAT92E. Cell Signal. 2005;18:819–829. [PubMed: 16129580]
- 35.
- Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 2005;19(16):1861–70. [PMC free article: PMC1186186] [PubMed: 16055650]
- 36.
- Ghiglione C, Devergne O, Georgenthum E. et al. The Drosophila cytokine receptor Domeless controls border cell migration and epithelial polarization during oogenesis. Development. 2002;129(23):5437–47. [PubMed: 12403714]
- 37.
- Müller P, Kuttenkeuler D, Gesellchen V. et al. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436(7052):871–5. [PubMed: 16094372]
- 38.
- Henriksen MA, Betz A, Fuccillo MV. et al. Negative regulation of STAT92E by an N-terminally truncated STAT protein derived from an alternative promoter site. Genes Dev. 2002;16(18):2379–89. [PMC free article: PMC187436] [PubMed: 12231627]
- 39.
- Shi S, Larson K, Guo D. et al. Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol. 2008;10(4):489–96. [PMC free article: PMC3083919] [PubMed: 18344984]
- 40.
- Karsten P, Hader S, Zeidler MP. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech Dev. 2002;117(1-2):343–6. [PubMed: 12204282]
- 41.
- Callus BA, Mathey-Prevot B. SOCS36E a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21(31):4812–21. [PubMed: 12101419]
- 42.
- Rawlings JS, Rennebeck G, Harrison SM. et al. Two Drosophila suppressors of cytokine signaling (SOCS) differentially regulate JAK and EGFR pathway activities. BMC Cell Biol. 2004;5(1):38. [PMC free article: PMC526380] [PubMed: 15488148]
- 43.
- Liu B, Liao J, Rao X. et al. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci USA. 1998;95(18):10626–31. [PMC free article: PMC27945] [PubMed: 9724754]
- 44.
- Chung CD, Liao J, Liu B. et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278(5344):1803–5. [PubMed: 9388184]
- 45.
- Mohr SE, Boswell RE. Zimp encodes a homologue of mouse Miz1 and PIAS3 and is an essential gene in Drosophila melanogaster. Gene. 1999;229(1-2):109–16. [PubMed: 10095110]
- 46.
- Betz A, Lampen N, Martinek S. et al. A Drosophila PIAS homologue negatively regulates stat92E. Proc Natl Acad Sci USA. 2001;98(17):9563–8. [PMC free article: PMC55492] [PubMed: 11504941]
- 47.
- Hari KL, Cook KR, Karpen GH. The Drosophila Su(var)2-10 locus regulates chromosome structure and function and encodes a member of the PIAS protein family. Genes Dev. 2001;15(11):1334–48. [PMC free article: PMC312712] [PubMed: 11390354]
- 48.
- Bach EA, Vincent S, Zeidler MP. et al. A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics. 2003;165(3):1149–66. [PMC free article: PMC1462825] [PubMed: 14668372]
- 49.
- Arbouzova NI, Bach EA, Zeidler MP. Ken and barbie selectively regulates the expression of a subset of Jak/STAT pathway target genes. Curr Biol. 2006;16(1):80–8. [PubMed: 16401426]
- 50.
- Lukacsovich T, Yuge K, Awano W. et al. The ken and barbie gene encoding a putative transcription factor with a BTB domain and three zinc finger motifs functions in terminalia development of Drosophila. Arch Insect Biochem Physiol. 2003;54(2):77–94. [PubMed: 14518006]
- 51.
- Harris MB, Mostecki J, Rothman PB. Repression of an interleukin-4-responsive promoter requires cooperative BCL-6 function. J Biol Chem. 2005;280(13):13114–21. [PubMed: 15659391]
- 52.
- Hartatik T, Okada S, Okabe S. et al. Binding of BAZF and Bc16 to STAT6-binding DNA sequences. Biochem Biophys Res Commun. 2001;284(1):26–32. [PubMed: 11374866]
- 53.
- Morrison DK, Murakami MS, Cleghon V. Protein kinases and phosphatases in the Drosophila genome. J Cell Biol. 2000;150(2):F57–62. [PMC free article: PMC2180215] [PubMed: 10908587]
- 54.
- Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3(11):900–11. [PubMed: 14668806]
- 55.
- Chang YC, Lin SY, Liang SY. et al. Tyrosine phosphoproteomics and identification of substrates of protein tyrosine phosphatase dPTP61F in Drosophila S2 cells by mass spectrometry-based substrate trapping strategy. J Proteome Res. 2008;7(3):1055–66. [PubMed: 18281928]
- 56.
- Li J, Li W, Calhoun HC. et al. Patterns and functions of STAT activation during Drosophila embryogenesis. Mech Dev. 2003;120(12):1455–68. [PMC free article: PMC3090291] [PubMed: 14654218]
- 57.
- Lopes ES, Araujo HM. The maternal JAK/STAT pathway of Drosophila regulates embryonic dorsal-ventral patterning. Braz J Med Biol Res. 2004;37(12):1811–8. [PubMed: 15558187]
- 58.
- Jinks TM, Polydorides AD, Calhoun G. et al. The JAK/STAT signaling pathway is required for the initial choice of sexual identity in Drosophila melanogaster. Mol Cell. 2000;5(3):581–7. [PubMed: 10882142]
- 59.
- Sefton L, Timmer JR, Zhang Y. et al. An extracellular activator of the Drosophila JAK/STAT pathway is a sex-determination signal element. Nature. 2000;405(6789):970–3. [PubMed: 10879541]
- 60.
- Sanson B. Generating patterns from fields of cells. Examples from Drosophila segmentation. EMBO Rep. 2001;2(12):1083–8. [PMC free article: PMC1084173] [PubMed: 11743020]
- 61.
- Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev Biol. 1996;175(2):314–24. [PubMed: 8626035]
- 62.
- Isaac DD, Andrew DJ. Tubulogenesis in Drosophila: a requirement for the trachealess gene product. Genes Dev. 1996;10(1):103–17. [PubMed: 8557189]
- 63.
- Lengyel JA, Iwaki DD. It takes guts: the Drosophila hindgut as a model system for organogenesis. Dev Biol. 2002;243(1):1–19. [PubMed: 11846473]
- 64.
- Hoch M, Pankratz MJ. Control of gut development by fork head and cell signaling molecules in Drosophila. Mech Dev. 1996;58(1-2):3–14. [PubMed: 8887312]
- 65.
- Takashima SR. Murakami Regulation of pattern formation in the Drosophila hindgut by wg, hh, dpp and en. Mech Dev. 2001;101(1-2):79–90. [PubMed: 11231061]
- 66.
- Iwaki DD, Johansen KA, Singer JB. et al. Drumstick, bowl and lines are required for patterning and cell rearrangement in the Drosophila embryonic hindgut. Dev Biol. 2001;240(2):611–26. [PubMed: 11784087]
- 67.
- Johansen KA, Iwaki DD, Lengyel JA. Localized JAK/STAT signaling is required for oriented cell rearrangement in a tubular epithelium. Development. 2003;130(1):135–45. [PubMed: 12441298]
- 68.
- Pankratz MJ, Hoch M. Control of epithelial morphogenesis by cell signaling and integrin molecules in the Drosophila foregut. Development. 1995;121(6):1885–98. [PubMed: 7601002]
- 69.
- Fuss B, Hoch M. Drosophila endoderm development requires a novel homeobox gene which is a target of Wingless and Dpp signalling. Mech Dev. 1998;79(1-2):83–97. [PubMed: 10349623]
- 70.
- Fuss B, Josten F, Feix M. et al. Cell movements controlled by the Notch signalling cascade during foregut development in Drosophila. Development. 2004;131(7):1587–95. [PubMed: 14998929]
- 71.
- Tsai YC, Sun YH. Long-range effect of upd, a ligand for Jak/STAT pathway, on cell cycle in Drosophila eye development. Genesis. 2004;39(2):141–53. [PubMed: 15170700]
- 72.
- Mukherjee T, Schafer U, Zeidler MP. Identification of Drosophila genes modulating janus kinase/signal transducer and activator of transcription signal transduction. Genetics. 2006;172(3):1683–97. [PMC free article: PMC1456271] [PubMed: 16387886]
- 73.
- Mukherjee T, Hombria JC, Zeidler MP. Opposing roles for Drosophila JAK/STAT signalling during cellular proliferation. Oncogene. 2005;24(15):2503–11. [PubMed: 15735706]
- 74.
- Chen X, Oh SW, Zheng Z. et al. Cyclin D-Cdk4 and cyclin E-Cdk2 regulate the Jak/STAT signal transduction pathway in Drosophila. Dev Cell. 2003;4(2):179–90. [PubMed: 12586062]
- 75.
- Tsai YC, Yao JG, Chen PH. et al. Upd/Jak/STAT signaling represses wg transcription to allow initiation of morphogenetic furrow in Drosophila eye development. Dev Biol. 2007;306(2):760–71. [PubMed: 17498684]
- 76.
- Ekas LA, Baeg GH, Flaherty MS. et al. JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila. Development. 2006;133(23):4721–9. [PubMed: 17079268]
- 77.
- Zeidler MP, Perrimon N, Strutt DI. Polarity determination in the Drosophila eye: a novel role for unpaired and JAK/STAT signaling. Genes Dev. 1999;13(10):1342–53. [PMC free article: PMC316719] [PubMed: 10346822]
- 78.
- Zeidler MP, Perrimon N, Strutt DI. The four-jointed gene is required in the Drosophila eye for ommatidial polarity specification. Curr Biol. 1999;9(23):1363–72. [PubMed: 10607560]
- 79.
- Ayala-Camargo A, Ekas LA, Flaherty MS. et al. The JAK/STAT pathway regulates proximo-distal patterning in Drosophila. Dev Dyn. 2007;236(10):2721–30. [PubMed: 17626283]
- 80.
- Chao JL, Tsai YC, Chiu SJ. et al. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development. 2004;131(16):3839–47. [PubMed: 15253935]
- 81.
- Reynolds-Kenneally JM. Mlodzik Notch signaling controls proliferation through cell-autonomous and non-autonomous mechanisms in the Drosophila eye. Dev Biol. 2005;285(1):38–48. [PubMed: 16039641]
- 82.
- Assa-Kunik E, Torres IL, Schejter ED. et al. Drosophila follicle cells are patterned by multiple levels of Notch signaling and antagonism between the Notch and JAK/STAT pathways. Development. 2007;134(6):1161–9. [PubMed: 17332535]
- 83.
- Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev Cell. 2005;9(5):687–98. [PubMed: 16256743]
- 84.
- Herz HM, Chen Z, Scherr H. et al. vps25 mosaics display non-autonomous cell survival and overgrowth and autonomous apoptosis. Development. 2006;133(10):1871–80. [PMC free article: PMC2519036] [PubMed: 16611691]
- 85.
- Moberg KH, Schelble S, Burdick SK. et al. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit noncell-autonomous overgrowth. Dev Cell. 2005;9(5):699–710. [PubMed: 16256744]
- 86.
- Bach EA, Ekas LA, Ayala-Camargo A. et al. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7(3):323–31. [PubMed: 17008134]
- 87.
- Williams MJ. Drosophila hemopoiesis and cellular immunity. J Immunol. 2007;178(8):4711–6. [PubMed: 17404248]
- 88.
- Wood WA. Jacinto Drosophila melanogaster embryonic hemocytes: masters of multitasking. Nat Rev Mol Cell Biol. 2007;8(7):542–51. [PubMed: 17565363]
- 89.
- Sorrentino RP, Carton Y, Govind S. Cellular immune response to parasite infection in the Drosophila lymph gland is developmentally regulated. Dev Biol. 2002;243(1):65–80. [PubMed: 11846478]
- 90.
- Rizki TM, Rizki RM. Lamellocyte differentiation in Drosophila larvae parasitized by Leptopilina. Dev Comp Immunol. 1992;16(2-3):103–10. [PubMed: 1499832]
- 91.
- Lemaitre B, Meister M, Govind S. et al. Functional analysis and regulation of nuclear import of dorsal during the immune response in Drosophila. EMBO J. 1995;14(3):536–45. [PMC free article: PMC398111] [PubMed: 7859742]
- 92.
- Qiu P, Pan PC, Govind S. A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development. 1998;125(10):1909–20. [PubMed: 9550723]
- 93.
- Luo H, Rose PE, Roberts TM. et al. The Hopscotch Jak kinase requires the Raf pathway to promote blood cell activation and differentiation in Drosophila. Mol Genet Genomics. 2002;267(1):57–63. [PubMed: 11919715]
- 94.
- Mandal L, Martinez-Agosto JA, Evans CJ. et al. A Hedgehog- and Antennapedia-dependent niche maintains Drosophila haematopoietic precursors. Nature. 2007;446(7133):320–4. [PMC free article: PMC2807630] [PubMed: 17361183]
- 95.
- Duvic B, Hoffmann JA, Meister M. et al. Notch signaling controls lineage specification during Drosophila larval hematopoiesis. Curr Biol. 2002;12(22):1923–7. [PubMed: 12445385]
- 96.
- Hanratty WP, Ryerse JS. A genetic melanotic neoplasm of Drosophila melanogaster. Dev Biol. 1981;83(2):238–49. [PubMed: 6786941]
- 97.
- Lebestky T, Jung SH, Banerjee U. A Serrate-expressing signaling center controls Drosophila hematopoiesis. Genes Dev. 2003;17(3):348–53. [PMC free article: PMC195988] [PubMed: 12569125]
- 98.
- Jung SH, Evans CJ, Uemura C. et al. The Drosophila lymph gland as a developmental model of hematopoiesis. Development. 2005;132(11):2521–33. [PubMed: 15857916]
- 99.
- Krzemien J, Dubois L, Makki R. et al. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature. 2007;446(7133):325–8. [PubMed: 17361184]
- 100.
- Asha H, Nagy I, Kovacs G. et al. Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics. 2003;163(1):203–15. [PMC free article: PMC1462399] [PubMed: 12586708]
- 101.
- Kwon EJ, Park HS, Kim YS. et al. Transcriptional regulation of the Drosophila raf proto-oncogene by Drosophila STAT during development and in immune response. J Biol Chem. 2000;275(26):19824–30. [PubMed: 10764759]
- 102.
- Ferrandon D, Imler JL, Hetru C. et al. The Drosophila systemic immune response: sensing and signalling during bacterial and fungal infections. Nat Rev Immunol. 2007;7(11):862–74. [PubMed: 17948019]
- 103.
- Barillas-Mury C, Han YS, Seeley D. et al. Anopheles gambiae Ag-STAT, a new insect member of the STAT family, is activated in response to bacterial infection. Embo J. 1999;18(4):959–67. [PMC free article: PMC1171188] [PubMed: 10022838]
- 104.
- Lagueux M, Perrodou E, Levashina EA. et al. Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila. Proc Natl Acad Sci USA. 2000;97(21):11427–32. [PMC free article: PMC17216] [PubMed: 11027343]
- 105.
- Nonaka M, Azumi K, Ji X. et al. Opsonic complement component C3 in the solitary ascidian, Halocynthia roretzi. J Immunol. 1999;162(1):387–91. [PubMed: 9886411]
- 106.
- Boulay JL, O’Shea JJ, Paul WE. Molecular phylogeny within type I cytokines and their cognate receptors. Immunity. 2003;19(2):159–163. [PubMed: 12932349]
- 107.
- Ekengren S, Tryselius Y, Dushay MS. et al. A humoral stress response in Drosophila. Curr Biol. 2001;11(9):714–8. [PubMed: 11369236]
- 108.
- Ekengren S, Hultmark D. A family of Turandot-related genes in the humoral stress response of Drosophila. Biochem Biophys Res Commun. 2001;284(4):998–1003. [PubMed: 11409894]
- 109.
- Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev Cell. 2002;3(5):711–22. [PubMed: 12431377]
- 110.
- Dostert C, Jouanguy E, Irving P. et al. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol. 2005;6(9):946–53. [PubMed: 16086017]
- 111.
- Zhang J, Li L. Stem cell niche: microenvironment and beyond. J Biol Chem. 2008;283(15):9499–503. [PubMed: 18272517]
- 112.
- Jones DL, Wagers AJ. No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol. 2008;9(1):11–21. [PubMed: 18097443]
- 113.
- Wallenfang MR. Aging within the Stem Cell niche. Dev Cell. 2007;13(5):603–4. [PubMed: 17981128]
- 114.
- Li L, Xie T. Stem cell niche: structure and function. Annu Rev Cell Dev Biol. 2005;21:605–31. [PubMed: 16212509]
- 115.
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132(4):598–611. [PMC free article: PMC4505728] [PubMed: 18295578]
- 116.
- Shivdasani AA, Ingham PW. Regulation of stem cell maintenance and transit amplifying cell proliferation by tgf-beta signaling in Drosophila spermatogenesis. Curr Biol. 2003;13(23):2065–72. [PubMed: 14653996]
- 117.
- Kiger AA, Jones DL, Schulz C. et al. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294(5551):2542–5. [PubMed: 11752574]
- 118.
- Tulina N, Matunis E. Control of stem cell self-renewal in Drosophila spermatogenesis by JAK-STAT signaling. Science. 2001;294(5551):2546–9. [PubMed: 11752575]
- 119.
- Hardy RW, Tokuyasu KT, Lindsley DL. et al. The germinal proliferation center in the testis of Drosophila melanogaster. J Ultrastruct Res. 1979;69(2):180–90. [PubMed: 114676]
- 120.
- Gonczy P, DiNardo S. The germ line regulates somatic cyst cell proliferation and fate during Drosophila spermatogenesis. Development. 1996;122(8):2437–47. [PubMed: 8756289]
- 121.
- Kiger AA, White-Cooper H, Fuller MT. Somatic support cells restrict germline stem cell self-renewal and promote differentiation. Nature. 2000;407(6805):750–4. [PubMed: 11048722]
- 122.
- Tran J, Brenner TJ, DiNardo S. Somatic control over the germline stem cell lineage during Drosophila spermatogenesis. Nature. 2000;407(6805):754–7. [PubMed: 11048723]
- 123.
- Brawley C, Matunis E. Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science. 2004;304(5675):1331–4. [PubMed: 15143218]
- 124.
- Boyle M, Wong C, Rocha M. et al. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell. 2007;1(4):470–8. [PubMed: 18371382]
- 125.
- Kawase E, Wong MD, Ding BC. et al. Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development. 2004;131(6):1365–75. [PubMed: 14973292]
- 126.
- Xie TAC. Spradling decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell. 1998;94(2):251–60. [PubMed: 9695953]
- 127.
- Lopez-Onieva L, Fernandez-Minan A, Gonzalez-Reyes A. Jak/Stat signalling in niche support cells regulates dpp transcription to control germline stem cell maintenance in the Drosophila ovary. Development. 2008;135(3):533–40. [PubMed: 18171682]
- 128.
- Wang L, Li Z, Cai Y. The JAK/STAT pathway positively regulates DPP signaling in the Drosophila germline stem cell niche. J Cell Biol. 2008;180(4):721–8. [PMC free article: PMC2265565] [PubMed: 18283115]
- 129.
- Decotto E, Spradling AC. The Drosophila ovarian and testis stem cell niches: similar somatic stem cells and signals. Dev Cell. 2005;9(4):501–10. [PubMed: 16198292]
- 130.
- Singh SR, Liu W, Hou SX. The adult Drosophila malpighian tubules are maintained by multipotent stem cells. Cell Stem Cell. 2007;1(2):191–203. [PMC free article: PMC2040309] [PubMed: 18371350]
- 131.
- Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5(4):311–21. [PubMed: 15803157]
- 132.
- Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441(7097):1068–74. [PubMed: 16810241]
- 133.
- Constantinescu SN, Girardot M, Pecquet C. Mining for JAK-STAT mutations in cancer. Trends Biochem Sci. 2008;33(3):122–31. [PubMed: 18291658]
- 134.
- Silver DL, Montell DJ. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell. 2001;107(7):831–41. [PubMed: 11779460]
- 135.
- Ruohola H, Bremer KA, Baker D. et al. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell. 1991;66(3):433–49. [PubMed: 1907889]
- 136.
- Cummings CA, Cronmiller C. The daughterless gene functions together with Notch and Delta in the control of ovarian follicle development in Drosophila. Development. 1994;120(2):381–94. [PubMed: 8149916]
- 137.
- Lopez-Schier HD. St Johnston delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev. 2001;15(11):1393–405. [PMC free article: PMC312703] [PubMed: 11390359]
- 138.
- Baksa K, Parke T, Dobens LL. et al. The Drosophila STAT protein, stat92E, regulates follicle cell differentiation during oogenesis. Dev Biol. 2002;243(1):166–75. [PubMed: 11846485]
- 139.
- McGregor JR, Xi R, Harrison DA. JAK signaling is somatically required for follicle cell differentiation in Drosophila. Development. 2002;129(3):705–17. [PubMed: 11830571]
- 140.
- Beccari S, Teixeira L, Rorth P. The JAK/STAT pathway is required for border cell migration during Drosophila oogenesis. Mech Dev. 2002;111(1-2):115–23. [PubMed: 11804783]
- 141.
- Hou SX, Zheng Z, Chen X. et al. The Jak/STAT pathway in model organisms: emerging roles in cell movement. Dev Cell. 2002;3(6):765–78. [PubMed: 12479803]
- 142.
- Fire A, Xu S, Montgomery MK. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–11. [PubMed: 9486653]
- 143.
- Clemens JC, Worby CA, Simonson-Leff N. et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci USA. 2000;97(12):6499–503. [PMC free article: PMC18635] [PubMed: 10823906]
- 144.
- Boutros M, Kiger AA, Armknecht S. et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303(5659):832–835. [PubMed: 14764878]
- 145.
- Armknecht S, Boutros M, Kiger A. et al. High-throughput RNA interference screens in Drosophila tissue culture cells. Methods Enzymol. 2005;392:55–73. [PubMed: 15644175]
- 146.
- Perrimon N, Mathey-Prevot B. Applications of high-throughput RNA interference screens to problems in cell and developmental biology. Genetics. 2007;175(1):7–16. [PMC free article: PMC1775003] [PubMed: 17244760]
- 147.
- Mller P, Boutros M, Zeidler MP. Identification of JAK/STAT pathway regulators—insights from RNAi screens Sem Cell Dev Biol 200819(5)in press . [PMC free article: PMC2631610] [PubMed: 18586112]
- 148.
- Friedman A, Perrimon N. Genome-wide high-throughput screens in functional genomics. Curr Opin Genet Dev. 2004;14(5):470–6. [PubMed: 15380236]
- 149.
- Spring DR. Chemical genetics to chemical genomics: small molecules offer big insights. Chem Soc Rev. 2005;34(6):472–82. [PubMed: 16137160]
- 150.
- Perrimon N, Friedman A, Mathey-Prevot B. et al. Drug-target identification in Drosophila cells: combining high-throughout RNAi and small-molecule screens. Drug Discov Today. 2007;12(1-2):28–33. [PubMed: 17198970]
- 151.
- Jaenisch RA. Bird Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–54. [PubMed: 12610534]
- 152.
- Turner BM. Defining an epigenetic code. Nat Cell Biol. 2007;9(1):2–6. [PubMed: 17199124]
- 153.
- Zhang RP, Adams D. Heterochromatin and its relationship to cell senescence and cancer therapy. Cell Cycle. 2007;6(7):784–9. [PubMed: 17377503]
- 154.
- Champagne N, Pelletier N, Yang XJ. The monocytic leukemia zinc finger protein MOZ is a histone acetyltransferase. Oncogene. 2001;20(3):404–9. [PubMed: 11313971]
- 155.
- Badenhorst P, Voas M, Rebay I. et al. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 2002;16(24):3186–98. [PMC free article: PMC187504] [PubMed: 12502740]
- 156.
- Shi S, Calhoun HC, Xia F. et al. JAK signaling globally counteracts heterochromatic gene silencing. Nat Genet. 2006;38(9):1071–6. [PMC free article: PMC3092431] [PubMed: 16892059]
- 157.
- Kwon SY, Xiao H, Glover BP. et al. The nucleosome remodeling factor (NURF) regulates genes involved in Drosophila innate immunity. Dev Biol. 2008;316(2):538–47. [PubMed: 18334252]
- JAK/STAT Pathway Signalling in Drosophila Melanogaster - Madame Curie Bioscience...JAK/STAT Pathway Signalling in Drosophila Melanogaster - Madame Curie Bioscience Database
- Insulin-Like Growth Factors and Breast Cancer Therapy - Madame Curie Bioscience ...Insulin-Like Growth Factors and Breast Cancer Therapy - Madame Curie Bioscience Database
- Do DNA Triple Helices or Quadruplexes Have a Role in Transcription? - Madame Cur...Do DNA Triple Helices or Quadruplexes Have a Role in Transcription? - Madame Curie Bioscience Database
- Microarrays for Cancer Diagnosis and Classification - Madame Curie Bioscience Da...Microarrays for Cancer Diagnosis and Classification - Madame Curie Bioscience Database
- Regulation of the Rod Photoreceptor Cyclic Nucleotide-Gated Channel - Madame Cur...Regulation of the Rod Photoreceptor Cyclic Nucleotide-Gated Channel - Madame Curie Bioscience Database
Your browsing activity is empty.
Activity recording is turned off.
See more...