NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Featured Data: 2009 H1N1 Influenza Sequences
NCBI is the repository for the 2009 influenza virus sequences from the global H1N1 outbreak and is making every effort to make the sequences available as soon as possible. You can access the recent flu sequences and retrieve them individually from a special influenza virus resource page that is updated daily:
http://www.ncbi.nlm.nih.gov/genomes/FLU/SwineFlu.html
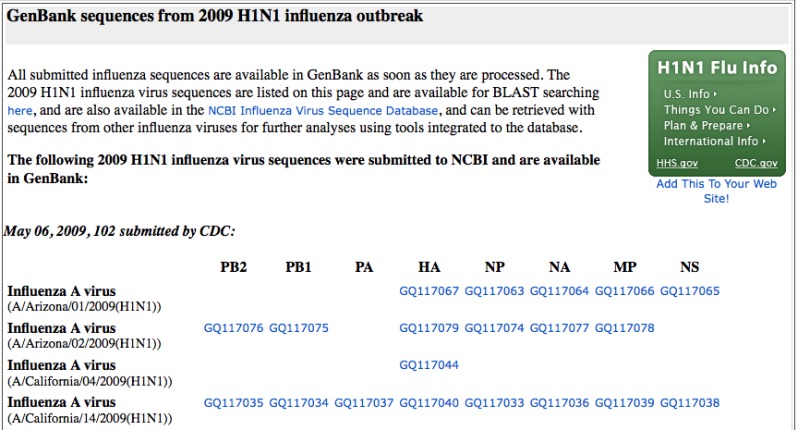
Using Flu Database Query Builder to Download Sequences in FASTA Format
An easy way to get these sequences all at once is through the query builder on the influenza virus database search page:
www.ncbi.nlm.nih.gov/genomes/FLU/Database/select.cgi
On this page you can select the characteristics of the flu sequences of interest and then retrieve them or perform additional analyses such as multiple alignments and phylogenetic tree construction.
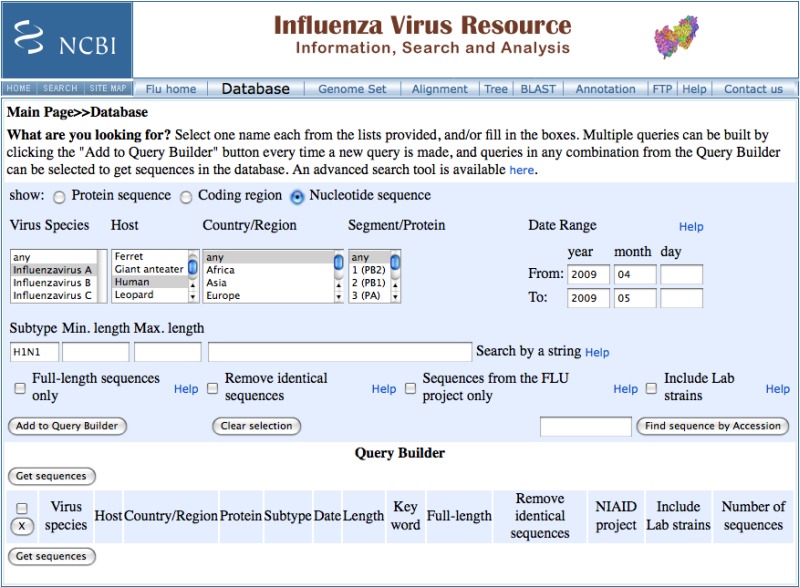
To get all of the recent H1N1 nucleotide sequences, select the nucleotide radio button on the “show” line of the form, set the species to “Influenzavirus A”, the host to “human”, the Country/Region to “any”, the collection date range to “2009/04:2009/05”, and the subtype to “H1N1”. Set the viral segment to “any” to get all segments, or select a specific segment or protein if desired. Click the “Get sequences” button to list the matching sequences. This search retrieves 304 records at the time of writing (May 7, 2009).
To download sequences in FASTA format choose the type of sequence desired – protein, coding region, nucleotide – from the “Select FASTA sequence for download” device at the top of the page. The file download dialog box appears that allows saving all sequences in FASTA format to a local file.
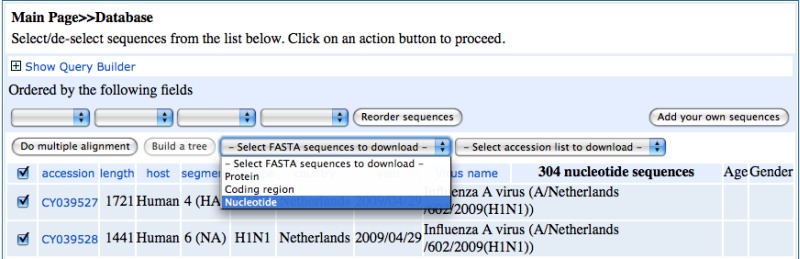
Using Batch Entrez to Download GenBank and Other Formats
Batch Entrez can be used to get the full records (GenBank, XML, or ASN.1) instead of the FASTA format for the flu sequences. To do this, download the list of accession numbers from the Flu database directly from the query builder results obtained above by selecting the desired sequence (protein or nucleotide) from the “Select accession list to download” device at the top of the results page. Save the list to a local file. Then upload these using the batch Entrez service to obtain the records as follows. Access the batch Entrez page.
www.ncbi.nlm.nih.gov/sites/batchentrez
Click the “Browse” button at the top of the batch Entrez page and point to the file containing the downloaded list of accessions. Click the “Retrieve” button then the link in the results to retrieve the influenza virus records in the Entrez nucleotide database. Once the records are in the Entrez Nucleotide service you can use the features of Entrez such as History and Preview/Index to refine your results if desired. See the Entrez help documentation for details.
www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=helpentrez
Click on individual records to in the results to view them in GenBank format or use the “Display” pull-down list to choose the format of interest and show all records. The records will be displayed 20 per page. Download the entire set by choosing the File option from the “Send to” pull-down list at the top of the first page of records.
Obtaining the eight diagnostic records
The World Health Organization has identified the eight segments of the earliest H1N1 isolate from California as diagnostic sequences for the new influenza virus strain.
www.who.int/csr/disease/swineflu/swineflu_genesequences_20090425.pdf
These sequences correspond to GenBank accession numbers FJ966079-FJ966086 available in the Entrez nucleotide service.
www.ncbi.nlm.nih.gov/sites/entrez?db=nuccore
Use the following query to retrieve these eight segments directly.
FJ966079:FJ966086[Accession]
Flu Sequence Updates
NCBI is expecting new data on a daily basis as the outbreak continues. Check the flu virus pages listed above for breaking news and new sequences.
Featured Resource: Protein Multiple Alignment Tool Web Service
NCBI will soon offer a Web multiple protein alignment service that uses the Constraint-Based Multiple Alignment Tool (COBALT)(1). COBALT can align a set of provided sequences or can be run as an extension to a Web BLAST search performing a multiple alignment on the set of protein sequences collected from the original BLAST search. The Web implementation of COBALT uses information from pairwise protein BLAST (blastp) scores, Conserved Domain Database results, and Prosite pattern matches as constraints in an initial pairwise alignment that is followed by a progressive multiple sequence alignment. The results from COBALT can be used with the BLAST treeview service to generate a phylogenetic tree from the multiple alignment. An often-requested service, the addition of a multiple alignment greatly enhances the suite of sequence analysis tools available at the NCBI and provides a new and powerful extension to BLAST.
Running COBALT on a Set of Sequences
The Web interface to COBALT will be available from the BLAST homepage or directly from the following URL:
www.ncbi.nlm.nih.gov/tools/cobalt/
The basic COBALT interface shown in Figure 1 A has the advanced parameters available through a link that expands the form. Advanced parameters include gap open and extend penalties and constraint and clustering parameters. The default values for the constraint and clustering parameters have been optimized to give the best alignment without undue sacrifice of speed. Generally, altering the default constraints will degrade performance. Detailed information on the any of the advanced parameters is available through help documentation linked to each option.
COBALT accepts protein sequences in FASTA format or NCBI identifiers as input. Figure 1 B shows a portion of the COBALT alignment obtained using nine of the protein sequences from the NCBI HomoloGene cluster for ATP citrate lyase (HomoloGene ID 854). The alignment contains an anomalous predicted protein from chimpanzee (XP_511495) that is mis-spliced because of missing data in the genome. This sequence creates large gaps in the multiple alignment that interrupt the conserved Citryl-Coa lyase domain. One very useful feature of the Web interface to COBALT is the ability to edit the sequence set and perform another alignment with the modified set (Figure 1 C). In this case, un-checking the box next to the chimpanzee sequence and re-submitting the set produces the improved alignment shown in Figure 1 D.
Running COBALT from Web BLAST Results
COBALT can also be run from the results of any Web protein BLAST search by clicking the “Multiple Alignment” link in the “Other reports” line on the BLAST results. This provides an easy way to collect homologs in a set of species and align them for phylogenetic or other comparative study. The most useful sets of sequences for these purposes come from searches with well-defined taxon-restricted databases. For example, a BLAST search with the human prolactin reference sequence (NP_000939) can collect growth hormone family members from bony fishes for building a multiple alignment and a gene tree. A BLAST search using the following settings finds 13 full-length growth hormone homologs from four species of fish: Database = Reference proteins (refseq_protein); Organism = bony fishes; Entrez query = srcdb refseq known[properties]; Expect threshold = 1e-6. The Entrez query limit eliminates proteins based entirely on gene predictions. The Expect threshold helps restrict the set to only closely related proteins and can be adjusted after expanding the “Algorithm parameters” of the BLAST form. A COBALT alignment can be generated by clicking the “Multiple alignment” link in the “Other reports” line at the top of the Descriptions or Alignments section of the BLAST results (Figure 2, top panel). The COBALT results appear in a new browser window when ready (Figure 2, middle panel).
Generating a Phylogenetic Tree from COBALT Results
A phylogenetic tree can be generated from any set of COBALT results by clicking the “Phylogenetic tree” link at the top (Figure 2, middle panel). The tree is generated using the Treeview (NCBI News, Summer 2006) feature of the BLAST Web service. This tree is calculated from a global multiple sequence alignment and is therefore more accurate than the Distance tree that can be generated from the BLAST results. The tree view display has options for redrawing the tree in different format, recalculating the distance metrics, downloading the tree in text formats, and displaying and realigning sequences from any node. The tree generated from a multiple alignment of the fish growth hormone family members collected by the BLAST search with the human prolactin precursor is shown on the bottom panel of Figure 2. The tree shows three distinct subfamilies: growth hormone, prolactin, and the somatolactins. The latter are pituitary hormones apparently found only in fishes(2).
Retrieving Previous COBALT Results
COBALT results from the Web service are stored at NCBI and are available for later retrieval in the same way as ordinary BLAST results. Recent COBALT results are available through the “Recent Results” tab at top of the COBALT submission or results pages. Like BLAST results COBALT results may be retrieved up 36 hours from the time of the search using the Request Identifier (RID) that uniquely identifies each set of results.
Summary and Future Directions
The COBALT multiple protein alignment tool expands the suite of sequence analysis tools available at the NCBI and provides a single pathway now for collecting related sequences using BLAST and then performing a rapid and accurate multiple alignment. Moreover for the first time multiple alignments can be used directly at the NCBI to generate and display phylogenetic trees making the NCBI Website a comprehensive resource for analyzing protein relationships. Upcoming improvements to the COBALT tool include the ability to re-format and download alignments in various standard formats such as FASTA plus gap. This will allow COBALT multiple alignments to be imported into other multiple alignmentprograms and editors.
New Databases and Tools
H1N1 Influenza Resources
NCBI has various resources available as described above. The Influenza Virus Resource has 34 H1N1 influenza sequences listed on the following page: www.ncbi.nlm.nih.gov/genomes/FLU/SwineFlu.html. PubMed contains recently added literature citations related to the new strain of influenza.
Peptide Data Resource
Peptidome is a new public repository that archives and distributes tandem mass spectrometry peptide and protein identification data. Web-based interfaces are available to browse and explore studies, peptides, and proteins. For more information see the Peptidome web page: www.ncbi.nlm.nih.gov/projects/peptidome/.
Genome Build
Build 1 of the Vitis vinifera (wine grape) genome is available in the Genomes database and on the NCBI Map Viewer. The Map Viewer page is: http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi?taxid=29760
Bookshelf
The Bookshelf has added a new chapter to the NCBI Help Manual, GaP FAQ Archive. The Bookshelf website URL is: www.ncbi.nlm.nih.gov/sites/entrez?db=Books
Microbial Genomes
Fifteen finished microbial genomes were released between March 24-April 29. The original sequence data files submitted to GenBank/EMBL/DDBJ are available on the FTP site: ftp.ncbi.nih.gov/genbank/genomes/Bacteria/. The RefSeq provisional versions of these genomes are also available: ftp.ncbi.nih.gov/genomes/Bacteria/.
GenBank News
GenBank release 171.0 is available via web and FTP. The current release includes information available as of April 10, 2009. Release notes are available on the on the NCBI ftp site: ftp.ncbi.nih.gov/genbank/gbrel.txt
NCBI is considering ceasing support for index files, so affected users are encouraged to read that section of the release notes and provide feedback to the GenBank group.
Updates and Enhancements
SRA Transcript BLAST
SRA transcript sequences are now searchable through a specialized BLAST page. All transcript sequences derived from 454 sequencing are available from NCBI’s SRA database. To perform a search, go to the SRA BLAST page.
GEO DataSet Browser
A new GEO DataSet Browser is available for browsing the curated gene expression DataSets. The new tool is located: www.ncbi.nlm.nih.gov/sites/GDSbrowser.
Sequence Analysis Tools Links in Entrez Sequence Databases
A new Sequence Analysis Tools section is available on the right hand Discovery column of Nucleotide and Protein records in the Entrez system. Sequence Analysis Tools contains links to the BLAST service for both protein and nucleotide sequences. Nucleotide records also link to Primer BLAST service. Both of these links load the currently viewed sequence in the submission area of the tool ready to perform a BLAST or Primer BLAST search. In addition protein records have a link to pre-computed conserved domain results. These new links make it easy to perform sequence analysis on the fly from any sequence record.
Exhibits
NCBI will be exhibiting at the American Society for Microbiology’s 190th General Meeting on May 17-21 in Philadelphia, Pennsylvania.
Announce Lists and RSS Feeds
Fifteen topic-specific mailing lists are described on the Announcement List summary page. Announce lists provide email announcements about changes and updates to NCBI resources. www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html
Seven RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce. Please see: www.ncbi.nlm.nih.gov/feed/
Comments and questions about NCBI resources may be sent to NCBI at: vog.hin.mln.ibcn@ofni, or by calling 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
References
- 1.
- Papadopoulos J, Agarwala R. COBALT: constraint-based alignment tool for multiple protein sequences. COBALT: constraint-based alignment tool for multiple protein sequences. 2007. [PubMed: 17332019]
- 2.
- Kaneko T. Cell biology of somatolactin. Cell biology of somatolactin. 1996. pp. 1–24. [PubMed: 8843651]
- PubMedLinks to PubMed
- NCBI News, May 2009 - NCBI NewsNCBI News, May 2009 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...