NCBI Bookshelf. A service of the National Library of Medicine, National Institutes of Health.
NCBI News [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 1991-2012.
Featured Resource: PubChem Now Offers 3-D Small Molecule Structures and a New Conformer Viewer (Pc3D)
NCBI’s PubChem now features calculated three-dimensional conformers (3-D conformers) for a large proportion of the PubChem compound database (17 million records, 88%). In addition, conformers are clustered to provide a list of similar 3-D conformers. These similar conformers provide a more relevant and expanded set of compounds with potentially similar biological and pharmacological activity. PubChem also provides a viewer for small molecule conformers and alignments that produces an animated view from displays in the Web service.
The new standalone viewer, Pc3D, is the small-molecule equivalent of the now standard Cn3D viewer for macromolecular structures and provides additional features for visualizing the details of small molecule structures and alignments. These powerful new aspects of PubChem greatly expand the potential of PubChem as a chemical informatics resource for rational biological inhibitor and drug design.
Conformers and similar conformers
Three–dimensional conformers and similarities in PubChem are calculated using the commercial software packages Omega and OEShape (OpenEye Scientific Software, Santa Fe, New Mexico) 1, 2. Conformers are provided for all molecules in PubChem that are not too large or complex. A single low-energy conformer is presented in the PubChem Web display and is used for the calculation of similar conformers. All PubChem data including conformers may be accessed through the NCBI Entrez system.
www.ncbi.nlm.nih.gov/sites/entrez?db=pccompound
These 3-D conformers are also available for download from the FTP site:
ftp.ncbi.nlm.nih.gov/pubchem/Compound_3D/
Conformer data is provided in NCBI Abstract Syntax Notation (ASN), Extensible Markup Language (XML), and the Structures Data File (SDF) format on both the Web service and the FTP site.
Similarity measurements incorporate both shape (Tanimoto shape) and feature similarity (Tanimoto color). Features include the presence of ring systems, hydrophobic entities, positive and negative ionizable groups, and hydrogen bond donors and acceptors. Similar conformers are defined as those with greater than 80% shape similarity and more than 50% feature similarity.
Similar conformers are available from the “Related Structures” link on the PubChem summaries in search results or from the Compound Information section of a PubChem record. The “View Conformers” link will load the aligned conformers directly into the Web-based viewer. Looking at similar conformers rather than the traditional 2-D similar compounds often can lead to additional related molecules with interesting properties. For example, the opiate morphine (Compound ID 5288826) currently has 1,297 similar compounds and 1,418 similar conformers in PubChem. The sets are quite different, however, with only 514 of the compounds common to both sets. The 3-D set in this case has distinct links to BioAssay and literature data. For other compounds there may be distinct links to protein structures as well. Thus examining the similar conformers can expand related compounds to a more diverse set of molecules.
Viewing small molecule conformers and alignments
Figure 1 shows a PubChem Compound record for the stimulant methylphenidate (Compound ID 4158). The small 2-D structural formula is the default graphic on the right-hand-side of the structure summary under the “Structure and Quick Link Bar”. Clicking on the “3D” tab produces the small 3-D graphic shown in the figure. The 3-D viewers are easily launched from this graphic.
Single conformers
A pop-up menu activated by clicking the 3-D graphic allows the conformer to be displayed in the Web-based viewer or in the standalone Pc3D viewer once it is installed. The standalone viewer is available for Windows, Linux and Mac OSX operating systems and may be downloaded and installed by following the instructions in the online Pc3D manual.
http://pubchem.ncbi.nlm.nih.gov/pc3d/
The top panel of figure 2 shows the single methylphenidate conformer displayed in the Web-based viewer. The conformer in the viewer is animated and rotates slightly about the three axes to provide perspective on the shape. Speed, zoom, rotation controls, and a toggle for hydrogen atoms on the left-hand-side of the viewer provide a means of changing the display. The Pc3D icon and link (“View in Pc3D”) within the Web-based viewer will download the structure and automatically display it in the installed Pc3D viewer. Pc3D has more flexible and sophisticated rendering options for conformers. The bottom panel of figure 2 shows methylphenidate rendered in a space-filling format, one of several display options available in Pc3D. The Options and Commands menus modify the display of the conformer. The online manual for Pc3D linked above has detailed instructions for using the various features of the program.
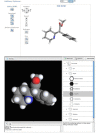
Figure 2
The lowest energy methylphenidate conformer rendered in the Web-based viewer (top panel) and in Pc3D. The style in Pc3D was changed to spacefill in this rendering.
Aligned conformers
Aligned conformers load directly into the Web-based viewer from the “View Conformers” link in the PubChem record as mentioned above. These alignments can also be loaded into an active view by clicking on the “Similar Conformers” link on the left-hand-side of the viewer. Pairwise conformer alignments are displayed one-at-a-time in the viewer. The alignment with the most similar conformer is shown first. Other alignments can be selected using the arrows at the top of the viewer to scroll through the alignments or by typing the number of the alignment in the collapsible search box beneath the arrows. The corresponding alignment can be loaded into the standalone viewer by clicking the “View in Pc3D” link. The alignment between methylphenidate and a pyrrolidine analog with inversion of configuration about the central carbon atom is shown in figure 3 for both the Web-based viewer (top panel) and Pc3D (bottom panel). (Note: The PubChem record for methylphenidate (CID 4158) does not specify the stereochemistry about the two chiral centers. In such cases, only a single enantiomer of the lowest energy structure is displayed and used for similarity calculations.)
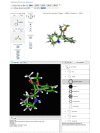
Figure 3
Alignment between a methylphenidate conformer and a conformer for a pyrrolidine analog that has inverted stereochemistry about the central carbon. Top panel, rendered in the Web-based viewer. Bottom panel, rendered in Pc3D.
Summary
The availability of 3-D conformers and similarities in PubChem greatly expands the utility of the PubChem resource by providing visualization tools and extends the universe of similar compounds. These now include not only simple derivatives but also additional compounds with similar volume, steric and charge features. Analyses using these features will help investigators understand the nature and causes of biological activity and provide new candidates for drug and inhibitor design.
References
1. Fontaine F, Bolton E, Borodina Y, Bryant SH. (2007) Fast 3D shape screening of large chemical databases through alignment-recycling. Chem Cent J. Jun 6;1:12.
2. Borodina YV, Bolton E, Fontaine F, Bryant SH. (2007) Assessment of conformational ensemble sizes necessary for specific resolutions of coverage of conformational space. J Chem Inf Model. Jul-Aug;47(4):1428-37.
New Databases and Tools
Genome Build
Build 1.1 of the genomes Hydra magnapapillata and Taeniopygia guttata (zebra finch) are available in the Genomes database and on the NCBI Map Viewer. The Map Viewer page is: http://www.ncbi.nlm.nih.gov/mapview/
Bookshelf
The Bookshelf has added four new books: Electrochemical Methods for Neuroscience, Frontiers in Neuroscience, Indwelling Neural Implants: Strategies for Contending with the InVivo Environment, and The National Academies Collection: Reports Funded by National Institutes of Health. The Bookshelf website URL is: www.ncbi.nlm.nih.gov/sites/entrez?db=Books
Microbial Genomes
Nine finished microbial genomes were released between February 7 and March 24. The original sequence data files submitted to GenBank/EMBL/DDBJ are available on the FTP site: ftp.ncbi.nih.gov/genbank/genomes/Bacteria/. The RefSeq provisional versions of these genomes are also available: ftp.ncbi.nih.gov/genomes/Bacteria/.
GenBank News
GenBank release 170.0 is available from the NCBI Web and FTP sites. The current release includes information available as of February 13, 2009. With this release, the new DBLINK linetype is now valid for GenBank sequence records, and it will begin to appear in GenBank Update files, soon after GenBank 170.0 is made available. Release notes are on the on the ftp site: ftp.ncbi.nih.gov/genbank/gbrel.txt
Updates and Enhancements
NCBI News
The NCBI News is now available in PDF format. There is a link for “Printable Copy” in the top right corner of each issue. Clicking that link will allow users to read and print a PDF version of the News.
RefSeq
RefSeq Release 34 is now available via Entrez and FTP. This full release incorporates genomic, transcript, and protein data available as of March 6, 2009. It includes 10,021,870 records from 8,054 different species and strains. The RefSeq website is: www.ncbi.nlm.nih.gov/RefSeq/. The FTP site is: ftp.ncbi.nlm.nih.gov/refseq/release. Changes since the previous release can be found in the release notes on the FTP site.
PubMed
Two new features have been added to the PubMed Abstract Plus Display to enhance discovery within the NCBI databases. Ads now appear in the right-hand discovery column, which provide additional links to related data. One new ad provides a link to the structure database if a structure is reported in the article, and a second ad provides links to PubMed Central articles that have cited the PubMed article being viewed.
Map Viewer
A new feature has been added to the Map Viewer homepage that links directly to a region on a chromosome. Clicking the “R” icon next to an organism name opens a box that provides options for chromosome number, assembly, and coordinate range. This feature links directly to the entered position on the given genome. The Map Viewer website is: www.ncbi.nlm.nih.gov/mapview/.
Exhibits
NCBI will be exhibiting at the American Society for Microbiology’s 190th General Meeting on May 17-21 in Philadelphia, Pennsylvania.
Announce Lists and RSS Feeds
Fifteen topic-specific mailing lists are described on the Announcement List summary page. Announce lists provide email announcements about changes and updates to NCBI resources. www.ncbi.nlm.nih.gov/Sitemap/Summary/email_lists.html
Seven RSS feeds are now available from NCBI including news on PubMed, PubMed Central, NCBI Bookshelf, LinkOut, HomoloGene, UniGene, and NCBI Announce. Please see: www.ncbi.nlm.nih.gov/feed/
Comments and questions about NCBI resources may be sent to NCBI at: vog.hin.mln.ibcn@ofni, or by calling 301-496-2475 between the hours of 8:30 a.m. and 5:30 p.m. EST, Monday through Friday.
- NCBI News, April 2009 - NCBI NewsNCBI News, April 2009 - NCBI News
Your browsing activity is empty.
Activity recording is turned off.
See more...